Abstract
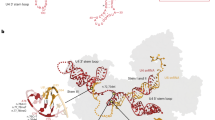
A narrow spectrum of heterozygous variants in RNU4-2, encoding the small nuclear RNA (snRNA) U4, underlies ReNU syndrome, a neurodevelopmental disorder (NDD) characterized by moderate to severe developmental delay (DD), intellectual disability (ID), a distinctive facial gestalt, and multisystem involvement. Pathogenic variants have primarily been reported within an 18-nt critical region contributing to stabilizing the U4/U6 snRNA duplex and proper spliceosome assembly. By combining whole genome sequencing reanalysis and targeted direct sequencing in 190 molecularly unexplained NDD cases, we report on five affected individuals carrying pathogenic/putative pathogenic RNU4-2 variants (2.6%). Three individuals harbored the recurrent pathogenic n.64_65insT variant, while two were heterozygous for private/rare maternally inherited variants (n.30 A > T and n.43_44insT) within the 5’ Stem-loop region. Deep clinical phenotyping confirmed a homogeneous constellation of features in all individuals, with global DD, ID, brain malformations, and a recognizable facial gestalt representing core findings. Based on structural homology models and available cryo-EM data, n.30 A > T and n.43_44insT were predicted to disrupt key intra- and inter-molecular interactions critical for spliceosome function. Our findings expand the mutational spectrum of ReNU syndrome, and confirm the 5’ Stem-loop as a second mutational hotspot in RNU4-2. We propose that a more complex genetics likely underlies the inheritance of a subset of disease-causing RNU4-2 variants from an apparently unaffected parent. We anticipate a relatively high proportion of pathogenic RNU4-2 variants among individuals with unclassified NDD despite extensive genomic testing, and propose a set of facial gestalt core features as a clinical screening tool to prioritize patients for RNU4-2 analysis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The sequencing data that support the findings of this work are available on request from the corresponding author. The data are not publicly available due to due to privacy/ethical restrictions. The new variants identified in this work and their clinical association have been submitted to ClinVar (n.30 A > T, VCV003390371.1; n.43_44insT, VCV003390370.1).
References
Alvarez-Mora MI, Sanchez A, Rodriguez-Revenga L, Corominas J, Rabionet R, Puig S, et al. Diagnostic yield of next-generation sequencing in 87 families with neurodevelopmental disorders. Orphanet J Rare Dis. 2022;17:60.
Wright CF, Campbell P, Eberhardt RY, Aitken S, Perrett D, Brent S, et al. Genomic diagnosis of rare pediatric disease in the United Kingdom and Ireland. N. Engl J Med. 2023;388:1559–71.
Chen Y, Dawes R, Kim HC, Ljungdahl A, Stenton SL, Walker S, et al. De novo variants in the RNU4-2 snRNA cause a frequent neurodevelopmental syndrome. Nature 2024;632:832–40.
Greene D, Thys C, Berry IR, Jarvis J, Ortibus E, Mumford AD, et al. Mutations in the U4 snRNA gene RNU4-2 cause one of the most prevalent monogenic neurodevelopmental disorders. Nat Med. 2024;30:2165–9.
Barbour K, Bainbridge MN, Wigby K, Besterman AD, Chuang NA, Tobin LE, et al. The face and features of RNU4-2: a new, common, recognizable, yet hidden neurodevelopmental disorder. Pediatr Neurol. 2024;161:188–93.
Burns VF, Radford EJ. ReNU syndrome - a newly discovered prevalent neurodevelopmental disorder. Trends Genet. 2024;40:914–6.
Wan R, Yan C, Bai R, Wang L, Huang M, Wong CC, et al. The 3.8A structure of the U4/U6.U5 tri-snRNP: Insights into spliceosome assembly and catalysis. Science (1979). 2016;351:466–75.
Nguyen THD, Galej WP, Bai X-C, Oubridge C, Newman AJ, Scheres SHW, et al. Cryo-EM structure of the yeast U4/U6.U5 tri-snRNP at 3.7 Å resolution. Nature 2016;530:298–302.
Nava C, Cogne B, Santini A, Leitão E, Lecoquierre F, Chen Y, et al. Dominant variants in major spliceosome U4 and U5 small nuclear RNA genes cause neurodevelopmental disorders through splicing disruption. medRxiv. 2024;:2024.10.07.24314689.
Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11 10 1–11 10 33.
DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–8.
Charenton C, Wilkinson ME, Nagai K. Mechanism of 5’ splice site transfer for human spliceosome activation. Science (1979). 2019;364:362–7.
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera-a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12.
Abramson J, Adler J, Dunger J, Evans R, Green T, Pritzel A, et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature. 2024;630:493–500.
Rosenblum J, Beysen D, Jansen AC, De Rademaeker M, Reyniers E, Janssens K, et al. RNU4-2-related neurodevelopmental disorder is associated with a recognisable facial gestalt. Clin Genet. 2025;107:104–12.
Schot R, Ferraro F, Geeven G, Diderich KEM, Barakat TS. Re-analysis of whole genome sequencing ends a diagnostic odyssey: Case report of an RNU4-2 related neurodevelopmental disorder. Clin Genet. 2024;106:512–7.
Ropers HH, Wienker T. Penetrance of pathogenic mutations in haploinsufficient genes for intellectual disability and related disorders. Eur J Med Genet. 2015;58:715–8.
Fernandes IR, Cruz ACP, Ferrasa A, Phan D, Herai RH, Muotri AR. Genetic variations on SETD5 underlying autistic conditions. Dev Neurobiol. 2018;78:500–18.
Powis Z, Farwell Hagman KD, Mroske C, McWalter K, Cohen JS, Colombo R, et al. Expansion and further delineation of the SETD5 phenotype leading to global developmental delay, variable dysmorphic features, and reduced penetrance. Clin Genet. 2018;93:752–761.
Amllal N, Elalaoui SC, Zerkaoui M, Chiguer A, Afif L, Izgua AT, et al. Identification of two novel ANKRD11 mutations: highlighting incomplete penetrance in KBG syndrome. Ann Lab Med. 2024;44:110–117.
Beunders G, Voorhoeve E, Golzio C, Pardo LM, Rosenfeld JA, Talkowski ME, et al. Exonic deletions in AUTS2 cause a syndromic form of intellectual disability and suggest a critical role for the C terminus. Am J Hum Genet. 2013;92:210–20.
Ansari M, Faour KNW, Shimamura A, Grimes G, Kao EM, Denhoff ER, et al. Heterozygous loss-of-function SMC3 variants are associated with variable growth and developmental features. HGG Adv. 2024;5:100273.
Greene D, De Wispelaere K, Lees J, Katrinecz A, Pascoal S, Hales E, et al. Mutations in the U2 snRNA gene RNU2-2P cause a severe neurodevelopmental disorder with prominent epilepsy. medRxiv. 2024:2024.09.03.24312863.
Quinodoz M, Rodenburg K, Cvackova Z, Kaminska K, de Bruijn SE, Iglesias-Romero AB, et al. De novo and inherited dominant variants in U4 and U6 snRNAs cause retinitis pigmentosa. medRxiv. 2025:2025.01.06.24317169.
van Rooij J, Arp P, Broer L, Verlouw J, van Rooij F, Kraaij R, et al. Reduced penetrance of pathogenic ACMG variants in a deeply phenotyped cohort study and evaluation of ClinVar classification over time. Genet Med. 2020;22:1812–1820.
Kingdom R, Wright CF. Incomplete penetrance and variable expressivity: from clinical studies to population cohorts. Front Genet. 2022;13:920390.
Cooper DN, Krawczak M, Polychronakos C, Tyler-Smith C, Kehrer-Sawatzki H. Where genotype is not predictive of phenotype: towards an understanding of the molecular basis of reduced penetrance in human inherited disease. Hum Genet. 2013;132:1077–130.
Stewart O, Gruber C, Randolph HE, et al. Monoallelic expression can govern penetrance of inborn errors of immunity. Nature 2025;637:1186–1197.
Acknowledgements
We wish to thank the families who participated in this study, and Prof. A Pichiecchio for the brain MRI data (Subjects 4 and 5). This work was performed in the frame of the collaborative research activity of the European Reference Network on Rare Congenital Malformations and Rare Intellectual Disability ERN-ITHACA (EU Framework Partnership Agreement ID: 3HP-HP-FPA ERN-01-2016/739516).
Funding
This work was supported, in part, by grants from the Italian Ministry of Health (Current Research Funds, to EMV and MT; RCR-2022-23682289 to FMS, EMV, GZ and MT; RF-2021-12374963, to MT).
Author information
Authors and Affiliations
Contributions
AB and MT conceived the work. AB, CMa, LC and MT wrote the manuscript. AB, CMa, LC, CC, AC, VC, and MF performed the genomic analyses, and analyzed and validated the genomic data. MC performed the structural analyses. MCB, SGC, ADF, LG, CL, CMe, RO, FP, DP, FSa, FSi, EMV and GZ collected the clinical data. MN, MP, and FCR performed the clinical data analyses. All coauthors provided critical feedback on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
The study was approved by the local Institutional Ethical Committee (ref. 1702_OPBG_2018 and 2072_OPBG_2020). Clinical data, pictures, and DNA samples were collected, used, and stored after signed informed consents from the participating subjects/families were secured. Written informed consents were obtained for publication of individual pictures
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Bruselles, A., Mancini, C., Chiriatti, L. et al. Expanding the mutational spectrum of ReNU syndrome: insights into 5’ Stem-loop variants. Eur J Hum Genet 33, 432–440 (2025). https://doi.org/10.1038/s41431-025-01820-1
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41431-025-01820-1