Abstract
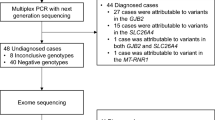
Hearing loss is a prevalent sensory disability with strong genetic heterogeneity, affecting approximately 60% of patients due to genetic factors. To investigate the possible genetic causes of hearing loss in 768 unrelated Chinese patients and analyze the genetic diagnosis rates among patients with different clinical phenotypic characteristics, 768 patients were enrolled, and whole-exome sequencing (WES) was performed for genetic testing. Sanger sequencing, MLPA, or qPCR were used to verify the parental origin of identified variants. We identified possible genetic etiologies in 501 of the 768 patients (65.2%), including 456 with non-syndromic and 45 with syndromic hearing loss. A total of 214 variants from 30 genes were identified: 174 previously reported and 40 novel pathogenic/likely pathogenic variants, the 18 hotspot variants accounted for 71.9% of all identified variants. Notably, 15 patients carried de novo variants. The genetic diagnosis rate was significantly higher in patients with severe-profound hearing loss (95.8%, 474/495) compared to those with mild-moderate hearing loss (9.9%, 27/273), P < 0.001. Our findings expand the spectrum of genetic variants associated with hearing loss and demonstrate high genetic diagnosis rates in patients with severe-profound hearing loss and congenital onset. WES combined with parental origin verification is an effective and economical method for identifying the genetic etiology of hearing loss and can be considered a priority in clinical practice for guiding early intervention and preventing further hearing loss.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request. The data are not publicly available due to privacy or ethical restrictions.
References
Sheffield AM, Smith RJH The Epidemiology of Deafness. Cold Spring Harbor perspectives in medicine 2019; 9.
Chadha S, Kamenov K, Cieza A. The world report on hearing, 2021. Bull World Health Organ. 2021;99:242–242a.
Writing Group For Practice Guidelines For D, Treatment Of Genetic Diseases Medical Genetics Branch Of Chinese Medical A, Yuan H, Dai P, Liu Y, Yang T. [Clinical practice guidelines for hereditary non-syndromic deafness]. Zhonghua yi xue yi chuan xue za zhi = Zhonghua yixue yichuanxue zazhi = Chin J Med Genet. 2020;37:269–76.
Shearer AE, Hildebrand MS, Sloan CM, Smith RJ. Deafness in the genomics era. Hearing Res. 2011;282:1–9.
Azaiez H, Booth KT, Ephraim SS, Crone B, Black-Ziegelbein EA, Marini RJ, et al. Smith RJH. Genomic Landscape and Mutational Signatures of Deafness-Associated Genes. Am J Hum Genet. 2018;103:484–97.
Shearer AE, DeLuca AP, Hildebrand MS, Taylor KR, Gurrola J 2nd, Scherer S, et al. Smith RJ. Comprehensive genetic testing for hereditary hearing loss using massively parallel sequencing. Proc Natl Acad Sci USA. 2010;107:21104–9.
Sloan-Heggen CM, Bierer AO, Shearer AE, Kolbe DL, Nishimura CJ, Frees KL, et al. Smith RJH. Comprehensive genetic testing in the clinical evaluation of 1119 patients with hearing loss. Hum Genet. 2016;135:441–50.
Pfundt R, Del Rosario M. Detection of clinically relevant copy-number variants by exome sequencing in a large cohort of genetic disorders. Genet Med. 2017;19:667–75.
Guan J, Li J, Chen G, Shi T, Lan L, Wu X, et al. Family trio-based sequencing in 404 sporadic bilateral hearing loss patients discovers recessive and De novo genetic variants in multiple ways. Eur J Med Genet. 2021;64:104311.
Pan J, Ma S, Teng Y, Liang D, Li Z, Wu L. Whole-exome sequencing identifies genetic variants of hearing loss in 113 Chinese families. Clin Chim Acta Int J Clin Chem. 2022;532:53–60.
Zeng B, Xu H, Yu Y, Li S, Tian Y, Li T, et al. Increased diagnostic yield in a cohort of hearing loss families using a comprehensive stepwise strategy of molecular testing. Front Genet. 2022;13:1057293.
Sun G, Huang W, Wang L, Wu J, Zhao G, Ren H, et al. Molecular findings in patients for whole exome sequencing and mitochondrial genome assessment. Clin Chim Acta. 2024;561:119774.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24.
Riggs ER, Andersen EF, Cherry AM, Kantarci S, Kearney H, Patel A, et al. Technical standards for the interpretation and reporting of constitutional copy-number variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen). Genet Med. 2020;22:245–57.
Kang H, Zhao K, Kong X. [Genetic testing of a Chinese pedigree affected with non-syndromic autosomal dominant deafness 15]. Zhonghua yi xue yi chuan xue za zhi = Zhonghua yixue yichuanxue zazhi = Chin J Med Genet. 2021;38:639–42.
Li J, Kang H, Kong X. Genetic diagnosis of Branchio-Oto syndrome pedigree due to a de novo heterozygous deletion of EYA1 gene. Zhonghua yi xue yi chuan xue za zhi= Zhonghua yixue yichuanxue zazhi= Chin J Med Genet. 2023;40:1128–33.
Zhao X, Chen C, Wei Y, Zhao G, Liu L, Wang C, et al. Novel mutations of COL4A3, COL4A4, and COL4A5 genes in Chinese patients with Alport Syndrome using next generation sequence technique. Mol Genet Genom Med. 2019;7:e653.
Shearer AE, Smith RJ. Massively Parallel Sequencing for Genetic Diagnosis of Hearing Loss: The New Standard of Care. Otolaryngol-Head Neck Surg. 2015;153:175–82.
Smith RJ, Bale JF Jr., White KR. Sensorineural hearing loss in children. Lancet. 2005;365:879–90.
Morton CC, Nance WE. Newborn hearing screening-a silent revolution. N Engl J Med. 2006;354:2151–64.
Shang H, Yan D, Tayebi N, Saeidi K, Sahebalzamani A, Feng Y, et al. Targeted Next-Generation Sequencing of a Deafness Gene Panel (MiamiOtoGenes) Analysis in Families Unsuitable for Linkage Analysis. Biomed Res Int. 2018;2018:3103986.
Wang Q, Xiang J, Sun J, Yang Y, Guan J, Wang D, et al. Nationwide population genetic screening improves outcomes of newborn screening for hearing loss in China. Genet Med. 2019;21:2231–8.
Kremer H. Hereditary hearing loss; about the known and the unknown. Hearing Res. 2019;376:58–68.
Korver AM, Smith RJ, Van Camp G, Schleiss MR, Bitner-Glindzicz MA, Lustig LR, et al. Congenital hearing loss. Nat Rev Dis Prim. 2017;3:16094.
Ling X, Wang C, Li L, Pan L, Huang C, Zhang C, et al. Third-generation sequencing for genetic disease. Clin Chim Acta Int J Clin Chem. 2023;551:117624.
Srivastava AK, Wang Y, Huang R, Skinner C, Thompson T, Pollard L, et al. Human genome meeting 2016: Houston, TX, USA. 28 February - 2 March 2016. Hum Genom. 2016;10:12.
Li J, Kang H, Kong X. [Diagnosis of a Chinese pedigree affected with autosomal recessive deafness 4 with enlarged vestibular aqueduct due to compound heterozygous variants of FOXI1 gene]. Zhonghua yi xue yi chuan xue za zhi = Zhonghua yixue yichuanxue zazhi = Chin J Med Genet. 2022;39:1080–4.
Acknowledgements
We thank the departmental staff for their assistance in the implementation of this work, and we are also very grateful to patients and their families included in the study for their support.
Funding
This work was supported by the National Key R&D Program of China (2018YFC1002206), Natural Science Foundation of Henan Province (232300420230, 242300420392) and Henan Provincial Key Scientific Research Project Plan of Higher Education Institution (25A310012).
Author information
Authors and Affiliations
Contributions
X Kong and H Kang conceived and designed the project. H Kang and J Li summarized all the data. H Kang, H Duan, Y Xia, L Su and Z Li performed genetic testing. H Kang and X Kong recruited patients. H Kang and J Li wrote the manuscript with help from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
This study was approved by the Medical Ethics Committee of the First Affiliated Hospital of Zhengzhou University (Approval no.:KS-2018-KY-36).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
41431_2025_1896_MOESM2_ESM.docx
Supplementary Table S2. The top 18 high frequency variants in all variant alleles (963) detected in 501 genetic positive cases
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kang, H., Li, J., Duan, H. et al. Clinical application of whole exome sequencing (WES) in the genetic diagnosis of 768 Chinese patients with bilateral hearing loss. Eur J Hum Genet (2025). https://doi.org/10.1038/s41431-025-01896-9
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41431-025-01896-9