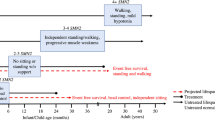
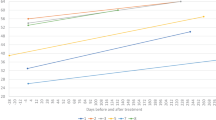
Abstract
Spinal muscular atrophy (SMA) is a progressive disease that affects motor neurons, with symptoms usually starting in infancy or early childhood. Recent breakthroughs in treatments targeting SMA have improved both lifespan and quality of life for infants and children with the disease. Given the impact of these treatments, it is essential to develop methods for managing treatment-induced changes in disease characteristics. Zolgensma® is the first effective and approved gene therapy for SMA caused by biallelic mutation in the SMN1 gene. In three children with SMA treated with Zolgensma®, neuronal, glial, inflammation, and vascular markers in the plasma exhibited a quicker response, emphasizing their potential as valuable biomarkers of treatment efficacy in clinical trials. We chose the novel Nucleic acid Linked Immuno-Sandwich Assay, to investigate a predefined panel of neuroinflammatory markers in plasma samples collected from SMA patients at baseline and six months after Zolgensma® treatment. We identified a set of novel targets whose levels differed between pre and post Zolgensma® treatment group and that were responsive to treatment. Even though our results warrant validation in larger SMA cohorts and longer follow-up time, they may pave the way for a panel of responsive proteins solidifying biomarker endpoints in SMA clinical trials.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 print issues and online access
$259.00 per year
only $43.17 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
Data used in this study are available within the article. Additional data supporting the information may be available upon reasonable request.
References
Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, Viollet L, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–65.
Arnold WD, Kassar D, Kissel JT. Spinal muscular atrophy: diagnosis and management in a new therapeutic era. Muscle Nerve. 2015;51:157–67.
Schroth M, Deans J, Arya K, Castro D, De Vivo DC, Gibbons MA, et al. Spinal Muscular Atrophy Update in Best Practices: Recommendations for Diagnosis Considerations. Neurol Clin Pr. 2024;14:e200310.
Darras BT, Masson R, Mazurkiewicz-Beldzinska M, Rose K, Xiong H, Zanoteli E, et al. Risdiplam-Treated Infants with Type 1 Spinal Muscular Atrophy versus Historical Controls. N Engl J Med. 2021;385:427–35.
Finkel RS, Mercuri E, Darras BT, Connolly AM, Kuntz NL, Kirschner J, et al. Nusinersen versus Sham Control in Infantile-Onset Spinal Muscular Atrophy. N Engl J Med. 2017;377:1723–32.
Mendell JR, Al-Zaidy S, Shell R, Arnold WD, Rodino-Klapac LR, Prior TW, et al. Single-Dose Gene-Replacement Therapy for Spinal Muscular Atrophy. N Engl J Med. 2017;377:1713–22.
Pino MG, Rich KA, Kolb SJ. Update on Biomarkers in Spinal Muscular Atrophy. Biomark Insights. 2021;16:11772719211035643.
Bayoumy S, Verberk IMW, Vermunt L, Willemse E, den Dulk B, van der Ploeg AT, et al. Neurofilament light protein as a biomarker for spinal muscular atrophy: a review and reference ranges. Clin Chem Lab Med. 2024;62:1252–65.
Glascock J, Darras BT, Crawford TO, Sumner CJ, Kolb SJ, DiDonato C, et al. Identifying Biomarkers of Spinal Muscular Atrophy for Further Development. J Neuromuscul Dis. 2023;10:937–54.
Kolb SJ, Kissel JT. Spinal Muscular Atrophy. Neurol Clin. 2015;33:831–46.
Farrar MA, Kiernan MC. The Genetics of Spinal Muscular Atrophy: Progress and Challenges. Neurotherapeutics. 2015;12:290–302.
Babic M, Banovic M, Berecic I, Banic T, Babic Leko M, Ulamec M, et al. Molecular Biomarkers for the Diagnosis, Prognosis, and Pharmacodynamics of Spinal Muscular Atrophy. J Clin Med. 2023;12:5060.
Strauss KA, Farrar MA, Muntoni F, Saito K, Mendell JR, Servais L, et al. Onasemnogene abeparvovec for presymptomatic infants with three copies of SMN2 at risk for spinal muscular atrophy: the Phase III SPR1NT trial. Nat Med. 2022;28:1390–7.
Chand D, Mohr F, McMillan H, Tukov FF, Montgomery K, Kleyn A, et al. Hepatotoxicity following administration of onasemnogene abeparvovec (AVXS-101) for the treatment of spinal muscular atrophy. J Hepatol. 2021;74:560–6.
Guillou J, de Pellegars A, Porcheret F, Fremeaux-Bacchi V, Allain-Launay E, Debord C, et al. Fatal thrombotic microangiopathy case following adeno-associated viral SMN gene therapy. Blood Adv. 2022;6:4266–70.
Galletta F, Cucinotta U, Marseglia L, Cacciola A, Gallizzi R, Cuzzocrea S, et al. Hemophagocytic lymphohistiocytosis following gene replacement therapy in a child with type 1 spinal muscular atrophy. J Clin Pharm Ther. 2022;47:1478–81.
Xie Q, Chen X, Ma H, Zhu Y, Ma Y, Jalinous L, et al. Improved gene therapy for spinal muscular atrophy in mice using codon-optimized hSMN1 transgene and hSMN1 gene-derived promotor. EMBO Mol Med. 2024;16:945–65.
Verma S, Perry K, Razdan R, Howell JC, Dawson AL, Hu WT. CSF IL-8 Associated with Response to Gene Therapy in a Case Series of Spinal Muscular Atrophy. Neurotherapeutics. 2023;20:245–53.
Feng W, Beer JC, Hao Q, Ariyapala IS, Sahajan A, Komarov A, et al. NULISA: a proteomic liquid biopsy platform with attomolar sensitivity and high multiplexing. Nat Commun. 2023;14:7238.
Bonanno S, Cavalcante P, Salvi E, Giagnorio E, Malacarne C, Cattaneo M, et al. Identification of a cytokine profile in serum and cerebrospinal fluid of pediatric and adult spinal muscular atrophy patients and its modulation upon nusinersen treatment. Front Cell Neurosci. 2022;16:982760.
Roberto J, Poulin KL, Parks RJ, Vacratsis PO. Label-free quantitative proteomic analysis of extracellular vesicles released from fibroblasts derived from patients with spinal muscular atrophy. Proteomics. 2021;21:e2000301.
Kolb SJ, Coffey CS, Yankey JW, Krosschell K, Arnold WD, Rutkove SB, et al. Baseline results of the NeuroNEXT spinal muscular atrophy infant biomarker study. Ann Clin Transl Neurol. 2016;3:132–45.
Kobayashi DT, Shi J, Stephen L, Ballard KL, Dewey R, Mapes J, et al. SMA-MAP: a plasma protein panel for spinal muscular atrophy. PLoS One. 2013;8:e60113.
Finkel RS, Crawford TO, Swoboda KJ, Kaufmann P, Juhasz P, Li X, et al. Candidate proteins, metabolites and transcripts in the Biomarkers for Spinal Muscular Atrophy (BforSMA) clinical study. PLoS One. 2012;7:e35462.
Niebroj-Dobosz I, Hausmanowa-Petrusewicz I. Serum cholinesterase activity in infantile and juvenile spinal muscular atrophy. Acta Neurol Scand. 1989;80:208–14.
Martinez-Hernandez R, Bernal S, Also-Rallo E, Alias L, Barcelo MJ, Hereu M, et al. Synaptic defects in type I spinal muscular atrophy in human development. J Pathol. 2013;229:49–61.
Harding BN, Kariya S, Monani UR, Chung WK, Benton M, Yum SW, et al. Spectrum of neuropathophysiology in spinal muscular atrophy type I. J Neuropathol Exp Neurol. 2015;74:15–24.
Yoshida M, Kitaoka S, Egawa N, Yamane M, Ikeda R, Tsukita K, et al. Modeling the early phenotype at the neuromuscular junction of spinal muscular atrophy using patient-derived iPSCs. Stem Cell Rep. 2015;4:561–8.
Murray LM, Comley LH, Thomson D, Parkinson N, Talbot K, Gillingwater TH. Selective vulnerability of motor neurons and dissociation of pre- and post-synaptic pathology at the neuromuscular junction in mouse models of spinal muscular atrophy. Hum Mol Genet. 2008;17:949–62.
Ling KK, Gibbs RM, Feng Z, Ko CP. Severe neuromuscular denervation of clinically relevant muscles in a mouse model of spinal muscular atrophy. Hum Mol Genet. 2012;21:185–95.
Kariya S, Park GH, Maeno-Hikichi Y, Leykekhman O, Lutz C, Arkovitz MS, et al. Reduced SMN protein impairs maturation of the neuromuscular junctions in mouse models of spinal muscular atrophy. Hum Mol Genet. 2008;17:2552–69.
Auer-Grumbach M, Toegel S, Schabhuttl M, Weinmann D, Chiari C, Bennett DLH, et al. Rare Variants in MME, Encoding Metalloprotease Neprilysin, Are Linked to Late-Onset Autosomal-Dominant Axonal Polyneuropathies. Am J Hum Genet. 2016;99:607–23.
Depondt C, Donatello S, Rai M, Wang FC, Manto M, Simonis N, et al. MME mutation in dominant spinocerebellar ataxia with neuropathy (SCA43). Neurol Genet. 2016;2:e94.
Higuchi Y, Hashiguchi A, Yuan J, Yoshimura A, Mitsui J, Ishiura H, et al. Mutations in MME cause an autosomal-recessive Charcot-Marie-Tooth disease type 2. Ann Neurol. 2016;79:659–72.
Scheijmans FEV, Cuppen I, Zwartkruis MM, Signoria I, van Ekris C, Asselman F, et al. Inflammatory markers in cerebrospinal fluid of paediatric spinal muscular atrophy patients receiving nusinersen treatment. Eur J Paediatr Neurol. 2023;42:34–41.
Nuzzo T, Russo R, Errico F, D’Amico A, Tewelde AG, Valletta M, et al. Nusinersen mitigates neuroinflammation in severe spinal muscular atrophy patients. Commun Med. 2023;3:28.
Acknowledgements
We thank patients for their donation of biofluids. We acknowledge the support of the coworkers of the Emory Health Sciences Research Building repository for sample storage and preparation. The authors would like to thank two anonymous reviewers for their constructive feedback. We would like to extend our gratitude to the Alamar TAP (technology access program) team for helpful discussion. DCP is supported by MDA DG and LLL CDA. This work was supported by the Children’s Healthcare of Atlanta 1998 Society Grant for SMA gene therapy to SV.
Author information
Authors and Affiliations
Contributions
DCP help wrote the draft and SV edited the manuscript. SV planned the study and provided access to the biofluids and de-identified individual participant data.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Pant, D.C., Verma, S. Identifying novel response markers for spinal muscular atrophy revealed by targeted proteomics following gene therapy. Gene Ther (2025). https://doi.org/10.1038/s41434-025-00513-0
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41434-025-00513-0