Abstract
Genetic/genomic manipulation techniques (gene transfer/delivery, gene editing, etc.) have become more and more mature, and the illegal use as gene doping in sports has drawn attentions. World Anti-Doping Agency (WADA) strictly prohibits gene doping, and has issued guideline on quantitative real-time PCR (qPCR) detections. However, the technical feature of qPCR makes it difficult to detect new doping targets, and codon changes on targets may also affect detection efficiency. Here, we prepare standard materials for genomic and transgenic versions of human EPO (hEPO) gene, and design qPCR primers to check the consequences of codon changes on gene doping detection. We confirm that carefully designed qPCR assays could indeed capture transgene signal, but codon changes on the transgene could severely undermine detection efficiency. We have also mimicked real world gene doping scenario by mixing genomic and transgenic versions of hEPO, and qPCR could detect wild-type but not codon-changed transgenes. As a method validation for such a challenge, we also use Sanger sequencing to confirm that sequencing could easily capture gene doping even for codon-changed transgenes. Our study confirms that codon changes will challenge qPCR-based gene doping detection, and calls for un-biased detection tools based on high-throughput sequencing in the future.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 print issues and online access
$259.00 per year
only $43.17 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All the data are provided in the manuscript.
References
Doudna JA. The promise and challenge of therapeutic genome editing. Nature. 2020;578:229–36.
Dunbar CE, High KA, Joung JK, Kohn DB, Ozawa K, Sadelain M. Gene therapy comes of age. Science. 2018;359:175.
Sun W, Wang H. Recent advances of genome editing and related technologies in China. Gene Ther. 2020;27:312–20.
Wong C. UK first to approve CRISPR treatment for diseases: what you need to know. Nature. 2023;623:676–7.
Reardon S. It’s a vote for hope’: first gene therapy for muscular dystrophy nears approval, but will it work?. Nature. 2023;618:451–3.
Lv J, Wang H, Cheng X, Chen Y, Wang D, Zhang L, et al. AAV1-hOTOF gene therapy for autosomal recessive deafness 9: a single-arm trial. Lancet. 2024;403:2317–25.
Musunuru K, Grandinette SA, Wang X, Hudson TR, Briseno K, Berry AM, et al. Patient-specific in vivo gene editing to treat a rare genetic disease. N Engl J Med. 2025;392:2235–43.
Filipp F. Is science killing sport? Gene therapy and its possible abuse in doping. Embo Rep. 2007;8:433–5.
Friedmann T, Rabin O, Frankel MS. Gene doping and sport. Science. 2010;327:647–8.
Pincock S. Gene doping. Lancet. 2005;366:S18–S9.
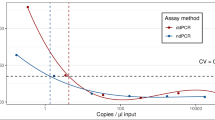
Li RH, Su PP, Shi Y, Shi H, Ding SQ, Su XB, et al. Gene doping detection in the era of genomics. Drug Test Anal. 2024;16:1468–78.
Lopez S, Meirelles J, Rayol V, Poralla G, Woldmar N, Fadel B, et al. Gene doping and genomic science in sports: where are we? Bioanalysis. 2020;12:801–11.
Gineviciene V, Utkus A, Pranckeviciene E, Semenova EA, Hall ECR, Ahmetov II. Perspectives in sports genomics. Biomedicines. 2022;10:298.
Baoutina A, Bhat S, Li DK, Emslie KR. Towards a robust test to detect gene doping for anabolic enhancement in human athletes. Drug Test Anal. 2023;15:314–23.
Yanazawa K, Sugasawa T, Aoki K, Nakano T, Kawakami Y, Takekoshi K. Development of a gene doping detection method to detect overexpressed human follistatin using an adenovirus vector in mice. PeerJ. 2021;9.e12285.
Kaiser J. How safe is a popular gene therapy vector? Science. 2020;367:131.
Smalley E. FDA warns public of dangers of DIY gene therapy. Nat Biotechnol. 2018;36:119–20.
WADA. The Prohibited List. https://www.wada-ama.org/en/prohibited-list. 2025.
Naumann N, Paßreiter A, Thomas A, Krug O, Walpurgis K, Thevis M. Analysis of potential gene doping preparations for transgenic DNA in the context of sports drug testing programs. Int J Mol Sci. 2023;24:15835.
Puchalska M, Witkowska-Piłaszewicz O. Gene doping in horse racing and equine sports: current landscape and future perspectives. Equine Vet J. 2025;57:312–24.
Tozaki T, Hamilton NA. Control of gene doping in human and horse sports. Gene Ther. 2022;29:107–12.
Paßreiter A, Thomas A, Grogna N, Delahaut P, Thevis M. First steps toward uncovering gene doping with CRISPR/Cas by identifying SpCas9 in plasma via HPLC–HRMS/MS. Anal Chem. 2020;92:16322–8.
Zheng B, Yan J, Li T, Zhao Y, Xu Z, Rao R, et al. Hydrophilic/hydrophobic modified microchip for detecting multiple gene doping candidates using CRISPR-Cas12a and RPA. Biosens Bioelectron. 2024;263:116631.
Paßreiter A, Naumann N, Thomas A, Grogna N, Delahaut P, Thevis M. Detection of sgRNA via SHERLOCK as potential CRISPR related gene doping control strategy. Anal Chem. 2024;96:7452–9.
Sugasawa T, Aoki K, Yanazawa K, Takekoshi K. Detection of multiple transgene fragments in a mouse model of gene doping based on plasmid vector using TaqMan-qPCR assay. Genes. 2020;11:750.
Neuberger EWI, Perez I, Le Guiner C, Moser D, Ehlert T, Allais M, et al. Establishment of two quantitative nested qPCR assays targeting the human EPO transgene. Gene Ther. 2016;23:330–9.
Tozaki T, Ohnuma A, Iwai S, Kikuchi M, Ishige T, Kakoi H, et al. Robustness of digital PCR and real-time PCR in transgene detection for gene-doping control. Anal Chem. 2021;93:7133–9.
Tozaki T, Ohnuma A, Kikuchi M, Ishige T, Kakoi H, Hirota KI, et al. Robustness of digital PCR and real-time PCR against inhibitors in transgene detection for gene doping control in equestrian sports. Drug Test Anal. 2021;13:1768–75.
Moser DA, Braga L, Raso A, Zacchigna S, Giacca M, Simon P. Transgene detection by digital droplet PCR. PLoS ONE. 2014;9:e111781.
Ohnuma A, Tozaki T, Kikuchi M, Ishige T, Kakoi H, Hirota K-i, et al. Multiplex detection of transgenes using πcode technology for gene doping control. Anal Chem. 2023;95:10149–54.
Salamin O, Kuuranne T, Saugy M, Leuenberger N. Loop-mediated isothermal amplification (LAMP) as an alternative to PCR: a rapid on-site detection of gene doping. Drug Test Anal. 2017;9:1731–7.
WADA. Laboratory Guidelines-Gene Doping Detection based on Polymerase Chain Reaction (PCR). https://www.wada-ama.org/en/resources/laboratory-guidelines-gene-doping-detection-based-polymerase-chain-reaction-pcr. 2021.
Naumann N, Do C, Vollmert C, Krajina M, Thomas A, Cheung HW, et al. Multiplex detection of seven transgenes for human gene doping analysis. Sci Rep. 2025;15:20219.
Wong KS, Cheung HW, Szeto CWL, Tsang CYN, Wan TSM, Ho ENM. A multiplex qPCR assay for transgenes detection: a novel approach for gene doping control in horseracing using conventional laboratory setup. Drug Test Anal. 2023;15:879–88.
Baoutina A, Bhat S, Zheng M, Partis L, Dobeson M, Alexander IE, et al. Synthetic certified DNA reference material for analysis of human erythropoietin transgene and transcript in gene doping and gene therapy. Gene Ther. 2016;23:708–17.
Baoutina A, Coldham T, Bains GS, Emslie KR. Gene doping detection: evaluation of approach for direct detection of gene transfer using erythropoietin as a model system. Gene Ther. 2010;17:1022–32.
Ostrander EA, Huson HJ, Ostrander GK. Genetics of athletic performance. Annu Rev Genomics Hum Genet. 2009;10:407–29.
Ren X, Shi Y, Xiao B, Su X, Shi H, He G, et al. Gene doping detection from the perspective of 3D genome. Drug Test Anal. 2025;17:1475–89.
Chin JX, Chung BKS, Lee DY. Codon Optimization OnLine (COOL): a web-based multi-objective optimization platform for synthetic gene design. Bioinformatics. 2014;30:2210–2.
Hernandez-Alias X, Benisty H, Radusky LG, Serrano L, Schaefer MH. Using protein-per-mRNA differences among human tissues in codon optimization. Genome Biol. 2023;24:34.
Thevis M, Kuuranne T, Geyer H. Annual banned-substance review 17th edition—analytical approaches in human sports drug testing 2023/2024. Drug Test Anal. 2025;17:1417–42.
Su X, Shi Y, Li R, Lu ZN, Zou X, Wu JX, et al. Application of qPCR assays based on haloacids transporter gene dehp2 for discrimination of Burkholderia and Paraburkholderia. BMC Microbiol. 2019;19:36.
Su X, Shi Y, Zou X, Lu Z-N, Xie G, Yang JYH, et al. Single-cell RNA-Seq analysis reveals dynamic trajectories during mouse liver development. BMC Genomics. 2017;18:946.
Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45.
van der Gronde T, de Hon O, Haisma HJ, Pieters T. Gene doping: an overview and current implications for athletes. Br J Sports Med. 2013;47:670–8.
Wells DJ. Gene doping: the hype and the reality. Br J Pharmacol. 2008;154:623–31.
Thompson H. Performance enhancement: superhuman athletes. Nature. 2012;487:287–9.
Bara S, Barczuk P, Gałajda E, Antonik J, Barczak I, Barć J, et al. A review of the possibilities of gene doping in sports focused on the advantages and disadvantages. J Educ Health Sport. 2023;42:35–45.
Baoutina A. A brief history of the development of a gene doping test. Bioanalysis. 2020;12:723–7.
Cantelmo RA, da Silva AP, Mendes-Junior CT, Dorta DJ. Gene doping: present and future. Eur J Sport Sci. 2020;20:1093–101.
Okano M, Ikekita A, Sato M, Inoue T, Kageyama S, Akiyama K, et al. Doping control analyses during the Tokyo 2020 Olympic and Paralympic Games. Drug Test Anal. 2022;14:1836–52.
Mauro VP, Chappell SA. A critical analysis of codon optimization in human therapeutics. Trends Mol Med. 2014;20:604–13.
Sugasawa T, Nakano T, Fujita S-i, Matsumoto Y, Ishihara G, Aoki K, et al. Proof of gene doping in a mouse model with a human erythropoietin gene transferred using an adenoviral vector. Genes. 2021;12:1249.
Beiter T, Zimmermann M, Fragasso A, Hudemann J, Niess AM, Bitzer M, et al. Direct and long-term detection of gene doping in conventional blood samples. Gene Ther. 2011;18:225–31.
Ni W, Le Guiner C, Gernoux G, Penaud-Budloo M, Moullier P, Snyder RO. Longevity of rAAV vector and plasmid DNA in blood after intramuscular injection in nonhuman primates: implications for gene doping. Gene Ther. 2011;18:709–18.
Lu Y, Yan J, Ou G, Fu L. A review of recent progress in drug doping and gene doping control analysis. Molecules. 2023;28:5483.
de Boer EN, van der Wouden PE, Johansson LF, van Diemen CC, Haisma HJ. A next-generation sequencing method for gene doping detection that distinguishes low levels of plasmid DNA against a background of genomic DNA. Gene Ther. 2019;26:338–46.
Tozaki T, Ohnuma A, Nakamura K, Hano K, Takasu M, Takahashi Y, et al. Detection of indiscriminate genetic manipulation in thoroughbred racehorses by targeted resequencing for gene-doping control. Genes. 2022;13:1589.
Tozaki T, Ohnuma A, Kikuchi M, Ishige T, Kakoi H, Hirota KI, et al. Whole-genome resequencing using genomic DNA extracted from horsehair roots for gene-doping control in horse sports. J Equine Sci. 2020;31:75–83.
Tozaki T, Ohnuma A, Takasu M, Nakamura K, Kikuchi M, Ishige T, et al. Detection of non-targeted transgenes by whole-genome resequencing for gene-doping control. Gene Ther. 2021;28:199–205.
Tozaki T, Ohnuma A, Kikuchi M, Ishige T, Kakoi H, Hirota K-i, et al. Rare and common variant discovery by whole-genome sequencing of 101 Thoroughbred racehorses. Sci Rep. 2021;11:16057.
Sutehall S, Malinsky F, Voss S, Chester N, Xu X, Pitsiladis Y. Practical steps to develop a transcriptomic test for blood doping. Transl Exerc Biomed. 2024;1:105–10.
Logsdon GA, Vollger MR, Eichler EE. Long-read human genome sequencing and its applications. Nat Rev Genet. 2020;21:597–614.
Maniego J, Giles O, Hincks P, Stewart G, Proudman C, Ryder E. Long-read sequencing assays designed to detect potential gene editing events in the myostatin gene revealed distinct haplotype signatures in the Thoroughbred horse population. Anim Genet. 2023;54:470–82.
Funding
This work is supported in part by Natural Science Foundation of Shanghai (25ZR1401192), Shanghai Gaofeng & Gaoyuan Project for University Academic Program Development (Shanghai University of Sport-2023), SJTU Interdisciplinary Program (YG2023QNB06), and Fundamental Research Funds for the Central Universities, Key Laboratory of Systems Biomedicine (Ministry of Education) Grant (KLSB2024QN-04).
Author information
Authors and Affiliations
Contributions
Conceptualization, original hypothesis, design of the study and methodology, XS; investigation, DW, SD, NL, YS1, PS, HS, YS2, BH, SC, XR, and FT; writing – original draft, RL; writing – review & editing, all authors; supervision, PC, JW, XS, and RL.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
Ethical approval was not needed for this paper as only synthesized DNA standard materials were used, without involvement of human or animal specimens/experiments.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wu, D., Ding, S., Liu, N. et al. Codon changes challenge PCR-based gene doping detection. Gene Ther (2025). https://doi.org/10.1038/s41434-025-00569-y
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41434-025-00569-y