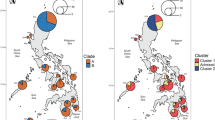
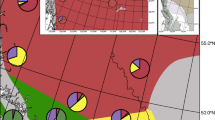
Abstract
Hybridization plays a major role in the evolutionary history of many taxa and can generate confounding patterns affecting many downstream applications. In this study, we empirically demonstrate how hybridization obfuscates phylogenetic inference (via the artefactual branch effect), species boundaries, and taxonomy in an adaptive radiation of frogs. Philippine narrow-mouthed frogs of the genus Kaloula exhibit a wide range of phenotypic and ecological adaptations but their evolutionary history and taxonomy remain poorly understood. In particular, the Kaloula conjuncta complex contains numerous subspecies with unresolved taxonomic boundaries and unclear evolutionary relationships. Within this complex, Kaloula conjuncta stickeli, until now was considered a rare, enigmatic, and phenotypically distinct subspecies that had not been encountered since its original description nearly 80 years ago. Here, we show that K. c. stickeli shares alleles with K. conjuncta meridionalis and another species outside the conjuncta group, K. picta. Using target-capture sequencing and a robust analytical framework, we show that despite having a unique phenotype, K. c. stickeli is likely an inviable F1 hybrid between K. c. meridionalis and K. picta and thus, does not warrant taxonomic recognition. Our results show how industry-standard approaches in systematic inference and integrative taxonomy—morphological, phylogenomic, clustering, and distance-based methods—can generate misleading results for identifying and understanding affinities of hybrids. In contrast, we demonstrate how network multispecies coalescent and population genetic approaches are more effective at accurately inferring reticulated evolutionary history. We also propose a rare phenomenon of deforestation-induced hybridization, which could have important consequences in light of large-scale Southeast Asian forest destruction.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Raw sequence reads have been deposited in the GenBank SRA as BioProject accession PRJNA1133022.
References
Abbott RJ, Barton NH, Good JM (2016) Genomics of hybridization and its evolutionary consequences. Mol Ecol 25:2325–2332
Alcala AC (1986) Guide to Philippine Flora and Fauna. Vol X, Amphibians and Reptiles. Natural Resource Management Center, Ministry of Natural Resource Management Center, Ministry of Natural Resources and the University of the Philippines, Manila, Philippines
Alcala AC, Brown RM (1998) Philippine Amphibians: An Illustrated Field Guide. Bookmark Press, Makati City, Philippines
Alexander AM, Su YC, Oliveros CH, Olson KV, Travers SL, Brown RM (2017) Genomic data reveals potential for hybridization, introgression, and incomplete lineage sorting to confound phylogenetic relationships in an adaptive radiation of narrow-mouth frogs. Evolution 71:475–488
Allman ES, Baños H, Rhodes JA (2019) NANUQ: A method for inferring species networks from gene trees under the coalescent model. Algorithms Mol Biol 14:1–25
Blackburn DC, Siler CD, Diesmos AC, Mcguire JA, Cannatella DC, Brown RM (2013) An adaptive radiation of frogs in a southeast Asian island archipelago. Evolution 67:2631–2646
Blischak PD, Chifman J, Wolfe AD, Kubatko LS (2018) HyDe: A python package for genome-scale hybridization detection. Syst Biol 67:821–829
Brown RM, Siler CD, Oliveros CH, Esselstyn JA, Diesmos AC, Hosner PA et al. (2013) Evolutionary processes of diversification in a model island archipelago. Annu Rev Ecol Evol Syst 44:411–435
Brown WC, Alcala AC (1970) The zoogeography of the herpetofauna of the Philippine Islands, a fringing archipelago. Proc Calif Acad Sci 38:105–130
Brown RM, Diesmos AC (2009). Philippines, Biology. In: Gillespie R, Clague D (eds) Encyclopedia of Islands, University of California Press: Berkeley, pp 723–732.
Burbrink FT, Guiher TJ (2015) Considering gene flow when using coalescent methods to delimit lineages of North American pitvipers of the genus Agkistrodon. Zool J Linn Soc 173:505–526
Burbrink FT, Crother BI, Murray CM, Smith BT, Ruane S, Myers EA et al. (2022) Empirical and philosophical problems with the subspecies rank. Ecol Evol 12:1–17
Burgon JD, Vences M, Steinfartz S, Bogaerts S, Bonato L, Donaire-Barroso D et al. (2021) Phylogenomic inference of species and subspecies diversity in the Palearctic salamander genus Salamandra. Mol Phylogenet Evol 157:107063
Catibog-Sinha CS, Heaney LR (2006) Philippine Biodiversity: principles and practice. Quezon City, Philippines, Haribon Foundation for Conservation of Natural Resources, pp 495
Chambers EA, Marshall TL, Hillis DM (2023) The Importance of Contact Zones for Distinguishing Interspecific from Intraspecific Geographic Variation. Syst Biol 72:357–371
Chan KO, Grismer LL (2021) A standardized and statistically defensible framework for quantitative morphological analyses in taxonomic studies. Zootaxa 5023:293–300
Chan KO, Grismer LL (2022) GroupStruct: An R package for allometric size correction. Zootaxa 5124:471–482
Chan KO, Mulcahy DG, Anuar S (2023) The artefactual branch effect and phylogenetic conflict: species delimitation with gene flow in mangrove pit vipers (Trimeresurus purpureomaculatus-erythrurus Complex). Syst Biol 72:1209–1219
Chan KO, Hutter CR, Wood PL, Grismer LL, Brown RM (2020a) Larger, unfiltered datasets are more effective at resolving phylogenetic conflict: Introns, exons, and UCEs resolve ambiguities in Golden-backed frogs (Anura: Ranidae; genus Hylarana). Mol Phylogenet Evol 151:106899
Chan KO, Hutter CR, Wood PLJ, Su Y-CC, Brown RM (2022) Gene flow increases phylogenetic structure and inflates cryptic species estimations: a case study on widespread Philippine Puddle Frogs (Occidozyga laevis). Syst Biol 71:40–57
Chan KO, Hutter CR, Wood PL, Grismer LL, Das I, Brown RM (2020b) Gene flow creates a mirage of cryptic species in a Southeast Asian spotted stream frog complex. Mol Ecol 29:3970–3987
Coates DJ, Byrne M, Moritz C (2018) Genetic diversity and conservation units: Dealing with the species-population continuum in the age of genomics. Front Ecol Evol 6:1–13
Darwin C (1859) On the origin of species by means of natural selection, or preservation of favoured races in the struggle for life. John Murray, London
De Queiroz K (2020) An updated concept of subspecies resolves a dispute about the taxonomy of incompletely separated lineages. Herpetol Rev 51:459–461
Derkarabetian S, Castillo S, Koo PK, Ovchinnikov S, Hedin M (2019) A demonstration of unsupervised machine learning in species delimitation. Mol Phylogenet Evol 139:106562
Diesmos A, Watters J, Huron N, Davis D, Alcala A, Crombie R et al. (2015) Amphibians of the Philippines, part I: checklist of the species. Proc Calif Acad Sci 62:457–539
Dolinay M, Nečas T, Zimkus BM, Schmitz A, Fokam EB, Lemmon EM et al. (2021) Gene flow in phylogenomics: Sequence capture resolves species limits and biogeography of Afromontane forest endemic frogs from the Cameroon Highlands. Mol Phylogenet Evol 163:107258
Durrant SD (1955) In defense of the subspecies. Syst Zool 4:186–190
Edelman NB, Mallet J (2021) Prevalence and Adaptive Impact of Introgression. Annu Rev Genet 55:265–283
Elgetany AH, van Rensburg H, Hektoen M, Matthee C, Budaeva N, Simon CA et al. (2020) Species delineation in the speciation grey zone—The case of Diopatra (Annelida, Onuphidae). Zool Scr 49:516–534
Environmental Center of the Philippines Foundation (1998) Environment and Natural Resources Atlas of the Philippines. Environmental Center of the Philippines Foundation, Manila, pp 394
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Frichot E, François O (2015) LEA: An R package for landscape and ecological association studies. Methods Ecol Evol 6:925–929
Frichot E, Mathieu F, Trouillon T, Bouchard G, François O (2014) Fast and efficient estimation of individual ancestry coefficients. Genetics 196:973–983
Frost DR (2024). Amphibian Species of the World: an Online Reference. Version 6.2 (last accessed 6 Sep 2024). Electronic Database accessible at http://research.amnh.org/herpetology/amphibia/index.html American Museum of Natural History, New York, USA.
Grant PR, Grant BR (2019) Hybridization increases population variation during adaptive radiation. Proc Natl Acad Sci USA 116:23216–23224
Handika H, Esselstyn JA (2024) SEGUL: Ultrafast, memory-efficient and mobile-friendly software for manipulating and summarizing phylogenomic datasets Mol Ecol Resour 24:e13964
Hillis D (2020) The detection and naming of geographic variation within species. Herpetol Rev 51(1):52–56
Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Le SV (2017) UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol 35:518–522
Huang JP, Knowles LL (2016) The species versus subspecies conundrum: Quantitative delimitation from integrating multiple data types within a single Bayesian approach in Hercules beetles. Syst Biol 65:685–699
Huson DH, Bryant D (2006) Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23:254–267
Hutter CR, Cobb KA, Portik DM, Travers SL, Brown RM, Wood PL et al. (2022) FrogCap: A modular sequence capture probe set for phylogenomics and population genetics for all frogs, assessed across multiple phylogenetic scales. Mol Ecol Resour 22:1100–1119
Inger RF (1954) Systematics and zoogeography of Philippine amphibia. Fieldiana 33:183–531
Kagawa K, Seehausen O (2020) The propagation of admixture-derived adaptive radiation potential. Proc Royal Soc B Biol Sci 287:20200941
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol 30:772–780
Krijthe JH (2015) Rtsne: T-Distributed Stochastic Neighbor Embedding using a Barnes-Hut Implementation. URL: https://github.com/jkrijthe/Rtsne.
Kummer DM (1992) Deforestation in the Postwar Philippines. Ateneo De Manila University Press, Manila, Philippines. pp 178
Lamb T, Avise JC (1986) Directional introgression of mitochondrial DNA in a hybrid population of tree frogs: The influence of mating behavior. Proc Natl Acad Sci USA 83:2526–2530
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N et al. (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079
Li W, Cerise JE, Yang Y, Han H (2017) Application of t-SNE to human genetic data. J Bioinf Comput Biol 15:1–14
Li H (2013). Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at https://doi.org/10.48550/arXiv.1303.3997.
Mallet J, Besansky N, Hahn MW (2016) How reticulated are species? BioEssays 38:140–149
Mallet J (2009) Rapid speciation, hybridization and adaptive radiation in the Heliconius melpomene group. In: Butlin RK, Bridle JR, Schluter D (eds) Speciation and Patterns of Diversity, Cambridge University Press, pp 177–194.
Mayr E (1982) Of what use are subspecies? Auk 99:593–595
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A et al. (2010) The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303
Mecham JS (1960) Introgressive hybridization between two southeastern treefrogs. Evolution 14:445–457
Meier JI, Marques DA, Mwaiko S, Wagner CE, Excoffier L, Seehausen O (2017) Ancient hybridization fuels rapid cichlid fish adaptive radiations. Nat Commun 8:14363
Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, Von Haeseler A et al. (2020) IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol 37:1530–1534
Moran BM, Payne C, Langdon Q, Powell DL, Brandvain Y, Schumer M (2021) The genomic consequences of hybridization. Elife 10:1–33
Patten MA (2015) Subspecies and the philosophy of science. Auk 132:481–485
Payseur BA, Rieseberg LH (2016) A genomic perspective on hybridization and speciation. Mol Ecol 25:2337–2360
Peñalba JV, Runemark A, Meier JI, Singh P, Wogan GOU, Sánchez-guillén R et al. (2024) The role of hybridization in species formation and persistence. Cold Spring Harb Perspect Biol 16:a041445
Pfeifer B, Wittelsbürger U, Ramos-Onsins SE, Lercher MJ (2014) PopGenome: An efficient Swiss army knife for population genomic analyses in R. Mol Biol Evol 31:1929–1936
Pyron RA, O’Connell KA, Duncan SC, Burbrink FT, Beamer DA (2023) Speciation hypotheses from phylogeographic delimitation yield an integrative taxonomy for seal salamanders (Desmognathus monticola). Syst Biol 72:179–197
R Core Team (2014) A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria
Roux C, Fraïsse C, Romiguier J, Anciaux Y, Galtier N, Bierne N (2016) Shedding light on the grey zone of speciation along a continuum of genomic divergence. PLoS Biol 14:1–22
Sackett LC, Seglund A, Guralnick RP, Mazzella MN, Wagner DM, Busch JD et al. (2014) Evidence for two subspecies of Gunnison’s prairie dogs (Cynomys gunnisoni), and the general importance of the subspecies concept. Biol Conserv 174:1–11
Sánchez KI, Huesa EGD, Breitman MF, Avila LJ, Sites JW, Morando M (2023) Complex patterns of diversification in the gray zone of speciation: model-based approaches applied to Patagonian liolaemid lizards (Squamata: Liolaemus kingii clade). Syst Biol 72:739–752
Sanguila MB, Cobb KA, Siler CD, Diesmos AC, Alcala AC, Brown RM (2016) The amphibians and reptiles of Mindanao Island, southern Philippines, II: The herpetofauna of northeast Mindanao and adjacent islands. Zookeys 624:1–132
Schiaffini MI (2020) Are subspecies (of Eira barbara) real? J Mammal 101:1410–1425
Seehausen O (2004) Hybridization and adaptive radiation. Trends Ecol Evol 19:198–207
Taylor SA, Larson EL (2019) Insights from genomes into the evolutionary importance and prevalence of hybridization in nature. Nat Ecol Evol 3:170–177
Thorpe RS (1983) A review of the numerical methods for recognising and analysing racial differentiation. In: Felsenstein J (ed) NATO ASI Series G1, Springer-Verlag: Berlin, Heidelberg, pp 404–423.
Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, del Angel G, Levy-Moonshine A et al. (2013) From fastQ data to high-confidence variant calls: The genome analysis toolkit best practices pipeline. Curr Protoc Bioinf 43:11.10.1–11.10.33
van der Maaten L, Hinton G (2008) Visualizing data using t-SNE. J Mach Learn Res 9:2579–2605
Vences M, Miralles A, Dufresnes C (2024) Next-generation species delimitation and taxonomy: Implications for biogeography. J Biogeogr 51:1709–1722
Wang J (2022) Fast and accurate population admixture inference from genotype data from a few microsatellites to millions of SNPs. Heredity 129:79–92
Wiens BJ, Colella JP (2024) triangulaR: an R package for identifying AIMs and building triangle plots using SNP data from hybrid zones. bioRxiv 03.28.5871.
Zhang C, Mirarab S (2022) Weighting by gene tree uncertainty improves accuracy of quartet-based species trees. Mol Biol Evol 39:1–22
Zhang C, Rabiee M, Sayyari E, Mirarab S (2018) ASTRAL-III: Polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinf 19:15–30
Acknowledgements
RMB acknowledges funding from the U. S. National Sciences Foundation for fieldwork (DEB 0073199 & 0743491) and development of genomic resources (DEB 1654388), and thanks the University of Kansas Office of the Provost Research Investment Council for the funding (2013–2014 R.I.C. Level II award 2300207) that initiated conversion of the KU Biodiversity Institute molecular genetics laboratory to collection & analysis of genome-scale data. KOC and PMH thank the University of Kansas Biodiversity Institute and Natural History Museum for postdoctoral fellowship support. RMB thanks Jim McGuire, Chris Austin, Isagani Bulalacao, Susing Babao, and Viscente Yngente for enthusiastic companionship during 1996 fieldwork on Mindanao Island, for sharing their own observations of frog behavior in multiple-species breeding aggregations, and for stimulating discussion regarding the possibility of interspecific hybridization among species of Philippine Kaloula.
Author information
Authors and Affiliations
Contributions
KOC, RMB, and PMH conceived the study and helped with writing. PMH collected genomic data and performed preliminary analyses. KOC conducted analyses and wrote the paper’s initial draft; all authors approved and contributed to the writing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Associate editor: Ben Evans.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chan, K.O., Hime, P.M. & Brown, R.M. Deforestation-induced Hybridization in Philippine Frogs Creates a Distinct Phenotype With an Inviable Genotype. Heredity 134, 200–208 (2025). https://doi.org/10.1038/s41437-025-00748-y
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41437-025-00748-y