Fig. 1: The parental dsDNA enters the zing finger region at the N-tier ring of CMG and is in-line with the unwound leading ssDNA.
From: DNA unwinding mechanism of a eukaryotic replicative CMG helicase
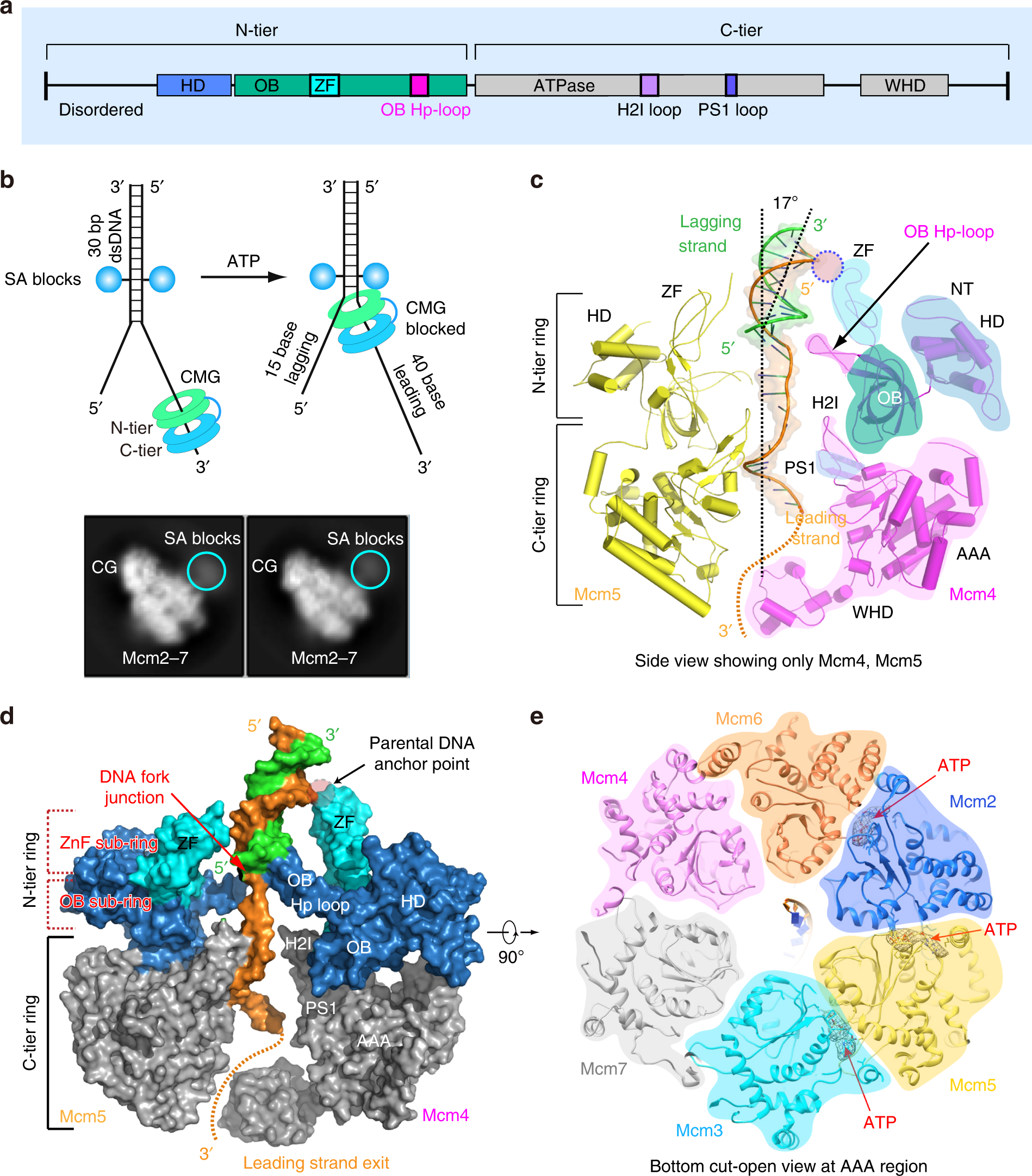
a A sketch of the general domain architecture of a typical Mcm protein, highlighting the hairpin loop in the OB domain (OB Hp-loop), and the H2I and PS1 loops in the AAA+ domain. HD: N-terminal helical domain, ZF: Zinc finger domain that is embedded in the OB domain, WHD: C-terminal winged helix domain. b Top panel: a sketch for the double streptavidin labeled forked DNA used to stall CMG in the presence of ATP. Bottom panel: two selected 2D class averages of the CMG stalled by the double streptavidin whose densities are visible. These averages are also shown in Fig. S1b. c, d Two side views of CMG–forked DNA complex in cartoon c and surface d views. Only Mcm4 and Mcm5 are shown to provide an unobstructed view of the forked DNA and the major structural features of a typical Mcm protein, as labeled in Mcm4. e A bottom view of Mcm2-7 that is cut open at the AAA+ motor ring. Cryo-EM densities for the three tentatively assigned ATP molecules are shown as gray mesh, superimposed with the atomic model of ATP in sticks.
