Fig. 1: Microbiota influences m6A modification in mouse cecum.
From: Impact of the gut microbiota on the m6A epitranscriptome of mouse cecum and liver
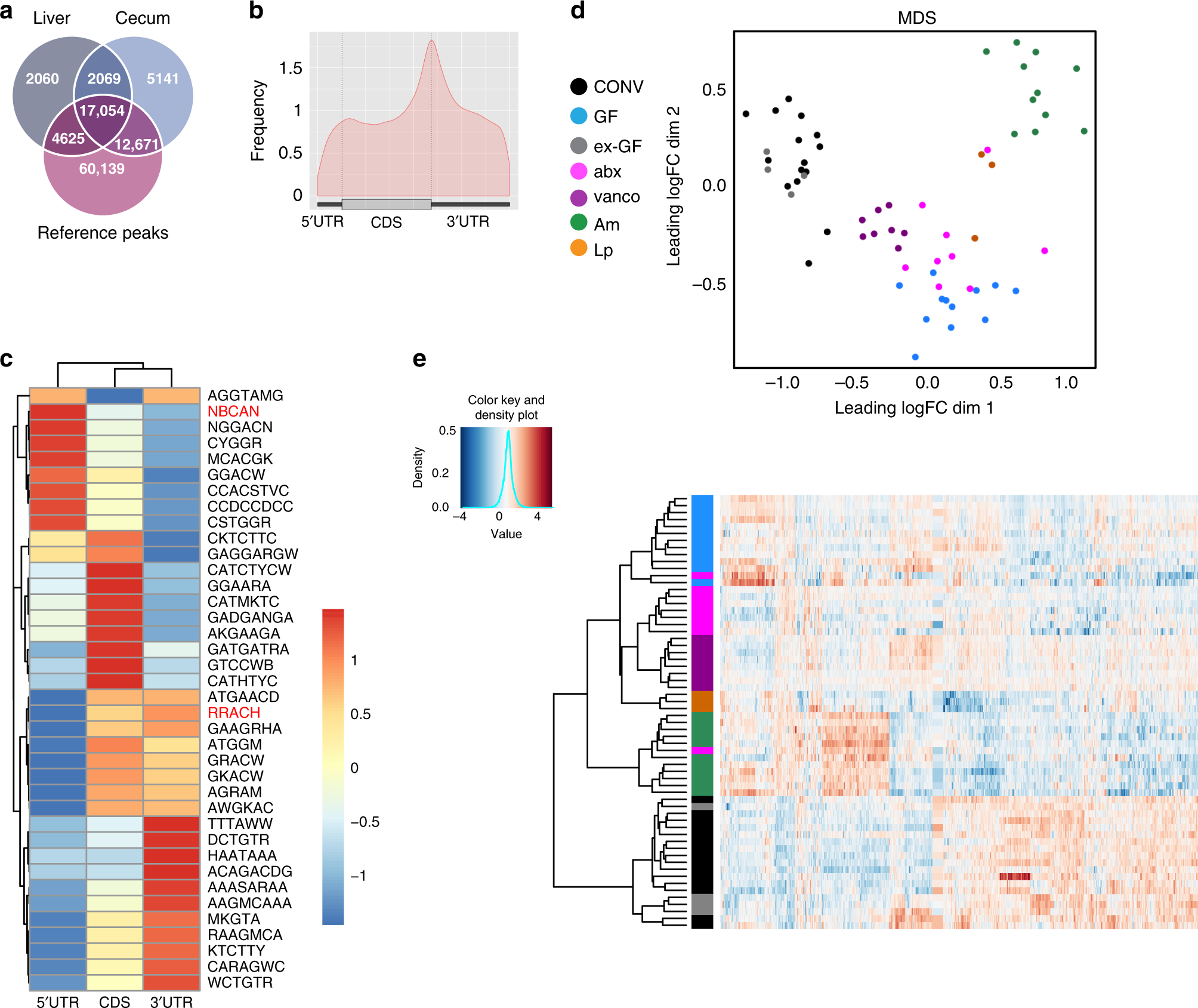
a 67% of m6A peaks in the cecum are overlapping with at least one liver peak, and 87% of detected peaks in the cecum are overlapping a MeT-DB v2.0 reference peak. For the liver tissue, 86% of the methylation peaks are overlapping at least one peak of the cecal tissue, and 89% are overlapping a MeT-DB v2.0 reference peaks. The reference peaks may be overlapping with several peaks we detected. b Positions of detected m6A peaks on all methylated transcripts in cecum were determined using the GUITAR package. c Motif enrichment in m6A modification was determined by calculating total occurrence of motifs in m6A peaks on the 5′UTR, CDS and 3′UTR of transcripts in the cecum. Consensus motifs for m6A (RRACH) and m6Am (NBCAN) are highlighted in red. d Multi-dimensional scaling (MDS) plot of the peak log2 counts-per-million IP data of all differentially methylated peaks showing the positions of the samples in the space spanned by the first and second MDS dimensions. Samples are colored with respect to condition: CONV (n = 15), conventionally raised mouse (black); GF (n = 12), germ-free mouse (cyan); ex-GF (n = 4), GF mouse colonized with the intestinal content of CONV mice (gray); abx (n = 9), CONV mice whose gut microbiota has been depleted by antibiotics treatment (magenta); vanco (n = 8), vancomycin/amphotericinB-treated mice (violet); Am (n = 11), A.muciniphila-mono-colonized mice (green), Lp (n = 3), L.plantarum-mono-colonized mice (orange); data were obtained from two independent sequencing experiments using ribodepleted RNA or purified mRNA from murine cecum. e Heat map of the peak log2 counts-per-million IP data of all differentially methylated peaks. Hierarchical clustering was performed using euclidean distance and ward.D2 linkage.
