Fig. 1: Design and validation of a yeast-display platform to identify peptide binding to a co-expressed class II MHC.
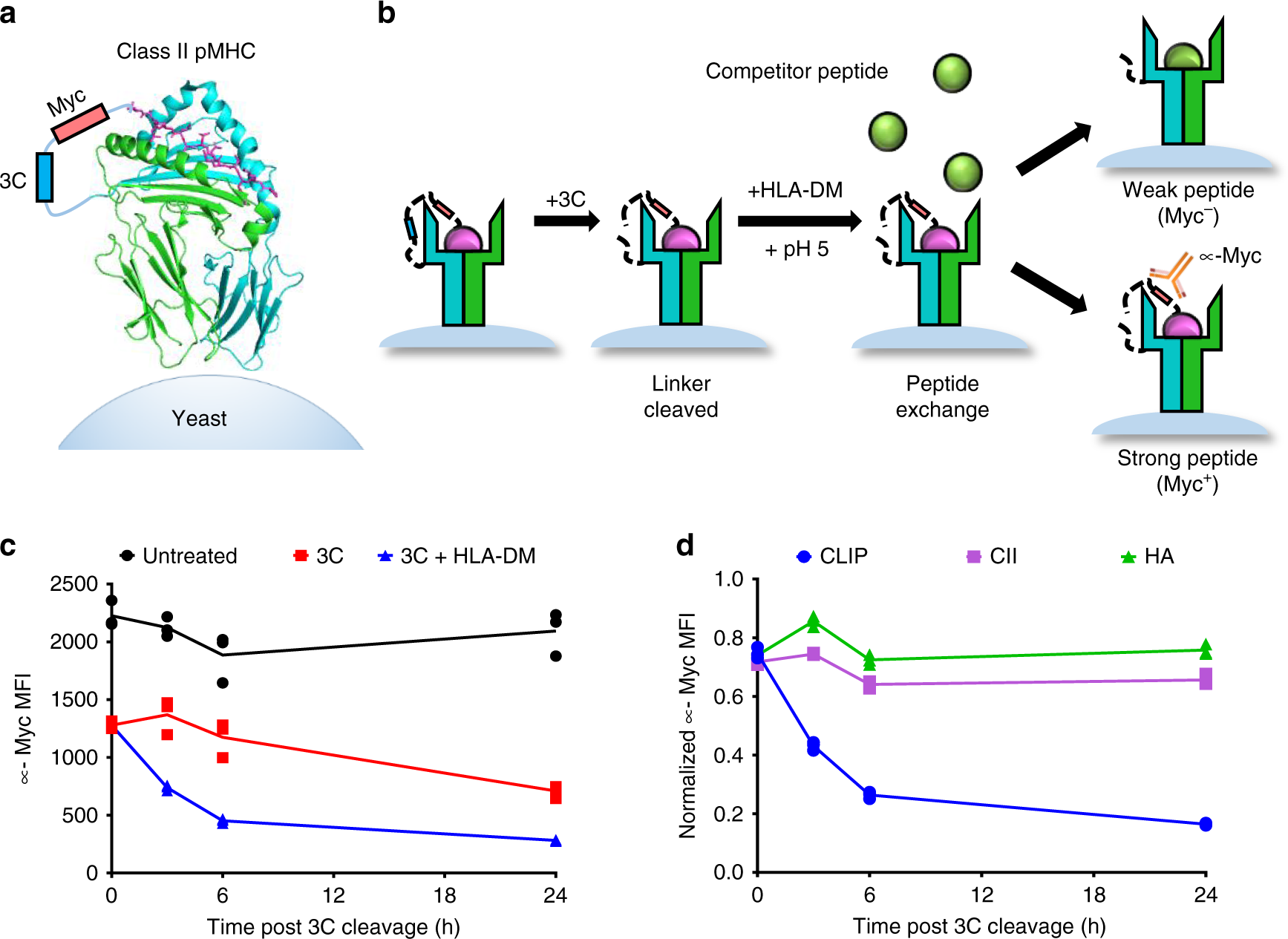
a Structural representation of HLA-DR401 (PDB 1J8H) modified to encode a 3C protease cleavage site and Myc epitope tag within the linker connecting the peptide and MHC β1 domain. b Schematic of validation protocol, including linker cleavage with 3C, peptide exchange at low pH in the presence of HLA-DM and high-affinity competitor peptide, and quantification of remaining bound peptide with an anti-Myc antibody. c Time course of mean fluorescence intensity (MFI) of a fluorescently labeled anti-Myc antibody for HLA-DR401-CLIP81-101-encoding yeast without treatment (Untreated), with linker cleavage (3C), or with linker cleavage and peptide exchange (3C + HLA-DM), as determined by flow cytometry. d Comparison of peptide retention for HLA-DR401-CLIP81-101, -CII261-273, or -HA306-318-encoding yeast with linker cleavage and peptide exchange, as determined by flow cytometry and normalized to MFI before treatment. For each construct, n = 3 aliquots were treated independently and measured for each time point and condition. Statistical evaluation was performed by repeated measures two-way ANOVA with Dunnett’s test for multiple comparison within treatment conditions (3 degrees of freedom, F = 54 in 1 C and F = 504 in 1D), or Tukey’s test for multiple comparisons across treatment conditions (2 degrees of freedom, F = 312 in 1 C and F = 2366 in 1D). Source data are provided as a Source Data file.
