Fig. 1: Deep learning frameworks are needed to accurately stratify toehold switches.
From: Sequence-to-function deep learning frameworks for engineered riboregulators
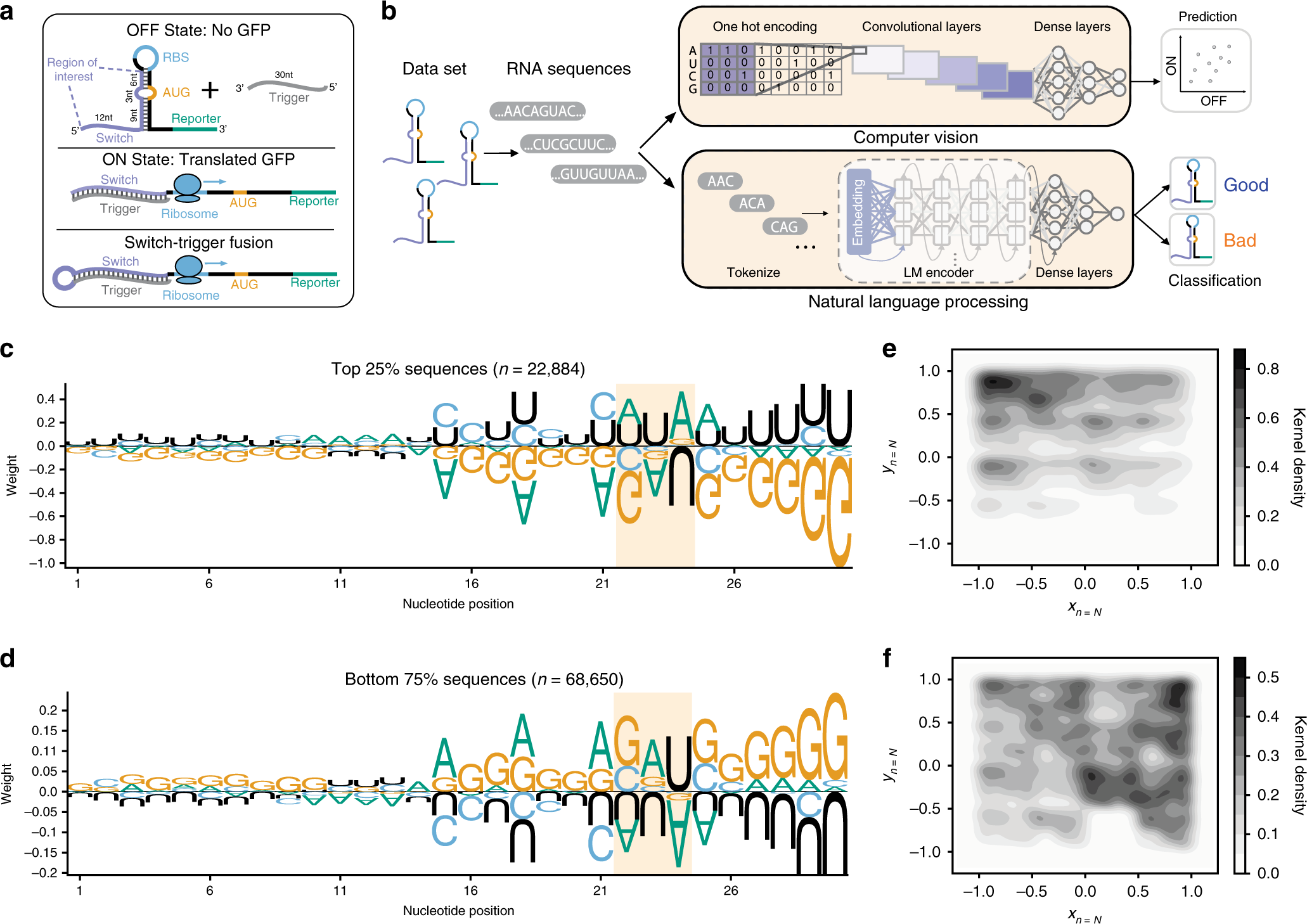
a Toehold switches modify their secondary structure in response to the presence of a complementary RNA molecule known as a trigger. In absence of the trigger, the Shine-Dalgarno sequence, or ribosome binding site (RBS), remains inaccessible and the reporter protein is not translated (OFF state). Upon binding of the trigger to the switch, the hairpin melts, allowing ribosome recruitment to the Shine–Dalgarno sequence and subsequent translation of the downstream reporter GFP protein (ON state). A modified ON state switch was built for experimental testing so that one molecule could be tested with trigger and switch fused together. b Two deep learning frameworks employing different strategies from computer vision and natural language processing were used to classify and predict toehold switch performance. c Sequence logos were calculated for the top 25% (N = 22,884) and d bottom 75% (N = 68,650) of sequences according to the experimental ON/OFF ratios. Weight corresponds to the log2 of each nucleotide probability normalized by the background frequency of that nucleotide in the set of all experimentally tested toeholds. Highlight indicates the motif found in positions 22–24 (Supplementary Fig. S1). e Chaos-game-representations (CGR) for the top 25% (N = 22,884) and f bottom 75% (N = 68,650) of toehold switches were calculated to visualize macroscopic sequence patterns, where darker regions correspond to an enrichment of sequences in that CGR locus. The sequence logos, CGR plots, and biophysical properties (Supplementary Fig. S3), however, were insufficient to characterize toehold performance on their own.
