Fig. 1: Illustrative overview of the workflow to create our Promoter transcription initiation frequency (TIF) Designer tool, ProD.
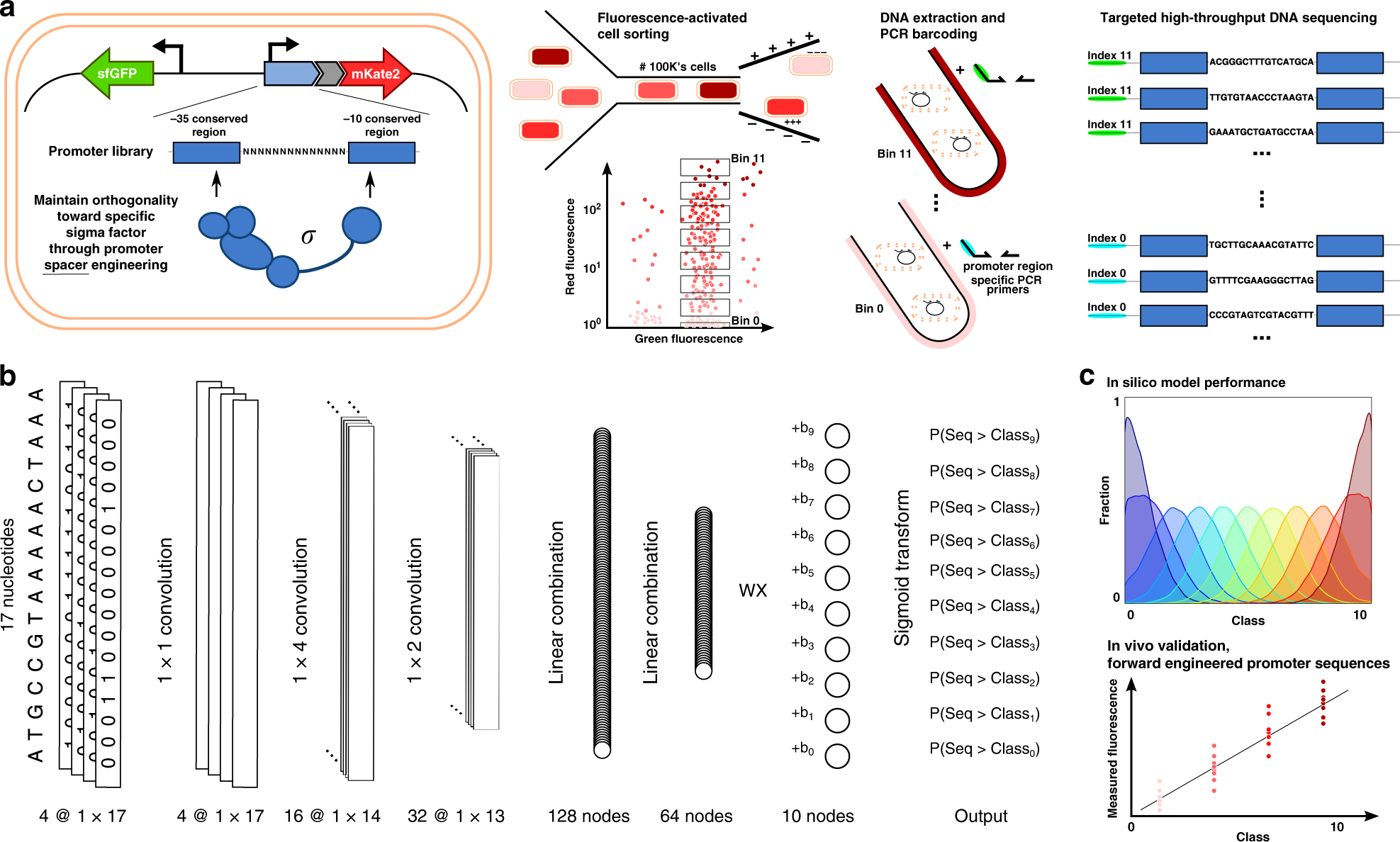
a Multiple promoter libraries were created, driving mKate2 expression, by engineering the promoter spacer DNA sequence to maintain the sigma factor recognition specificity. Cells were sorted by fluorescence-activated cell sorting in 12 separate bins, according to the level of red fluorescent protein expression. Subsequently, plasmid DNA was extracted and the promoter regions were amplified and barcoded uniquely for each bin. High-throughput DNA sequencing was used for genotyping. b The architecture of the neural network for ordinal regression trained on the created data sets for promoter TIF. Seventeen nucleotide sequences are processed by four 1 × 1, 16 1 × 4, and 32 1 × 2 convolutions and two fully connected layers of 128 and 64 nodes. A latent variable, correlated to the TIF of the promoter, is obtained through a single linear combination of weights (w) with the 64 output nodes (x). A vector of ordered biases b, optimized during training, outputs ten shifted values relative to the latent variable. The sigmoid transform of these outputs represents the probability of the TIF of the sequence being greater than a given class y. c The model with a minimum loss on the validation set is selected and evaluated on the test set, showing the ordinal correlation between its predictions and the true classes. By random sampling, a set of promoter sequences is generated and a selection is made, as predicted to display a range of promoter TIF levels covering the different classes (0–10), for in vivo validation (only performed for the E. coli σ70 promoter TIF model).
