Fig. 1: Quantifying off-target activity by CRISPECTOR.
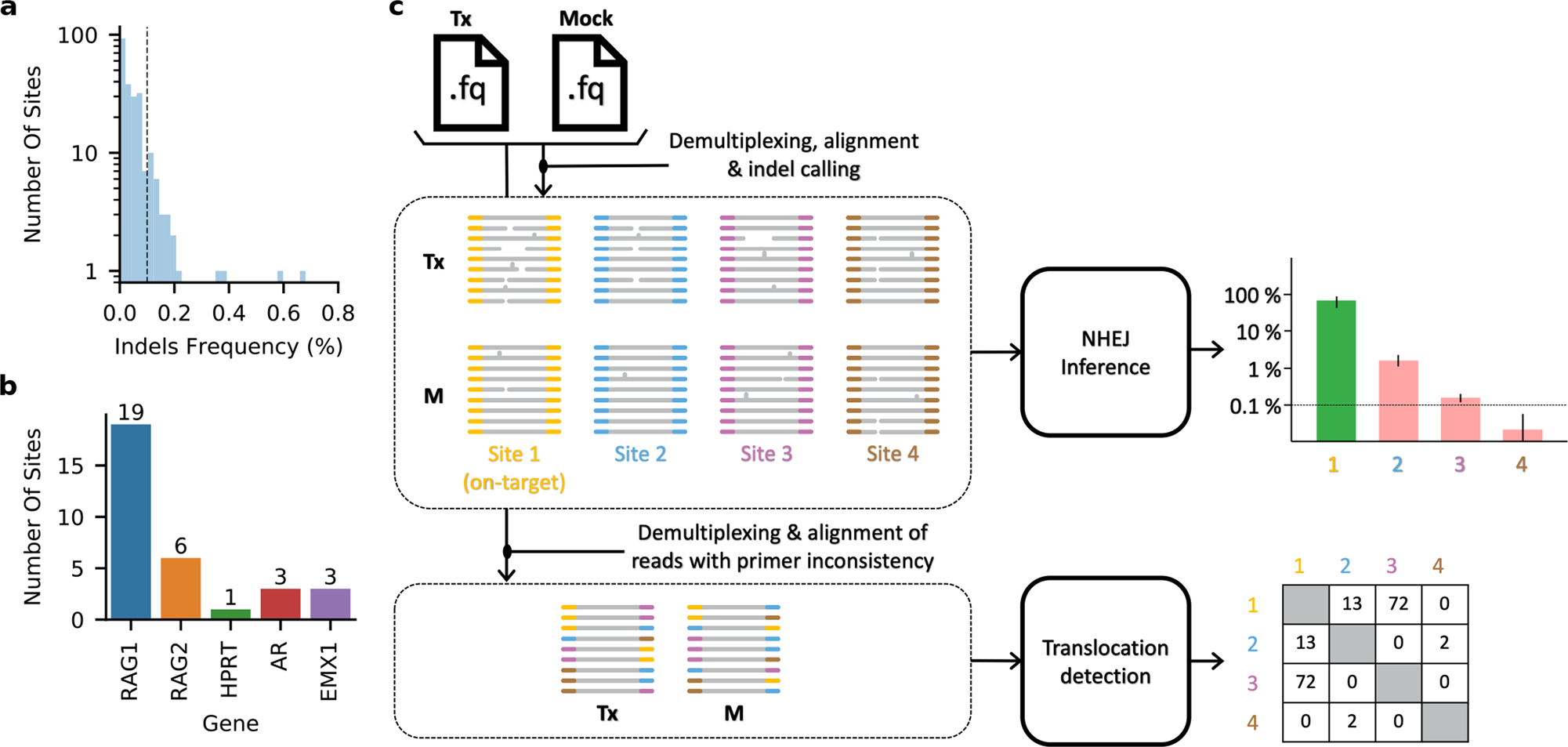
a, b Hypothetical indels at the expected cut-site in the M experiments. a Out of 226 off-target sites that we have examined in our experimental data, we have found 31 sites to have indel frequency higher than 0.1% (fraction = 0.001) in the M experiments. M indel frequencies at the cut-site were measured by a direct calculation, not using a comparative set-up. b These 31 noisy off-target sites come from different gRNAs examined, as depicted. (c) CRISPECTOR workflow. CRISPECTOR assigns each read in the Tx and M FASTQ files to a specific locus of interest or a putative translocation. Then, a Bayesian inference classifier accurately estimates the indel editing activity, and a hypergeometric test is performed to detect translocation reads.
