Fig. 1: In vitro RNase activity assays with the endogenous and reconstituted TtCmr complexes.
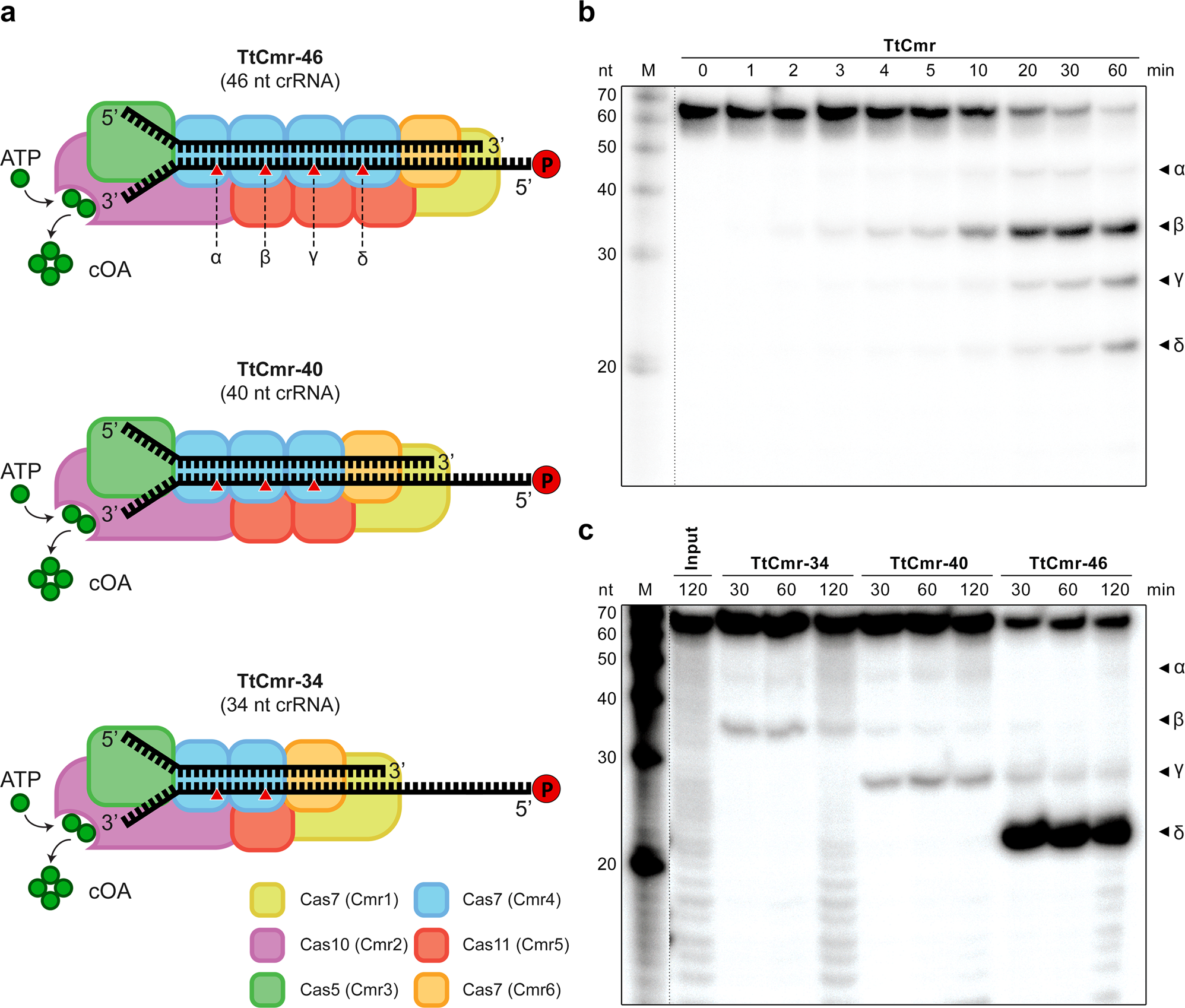
a Schematic illustration of the different reconstituted TtCmr complexes used in the activity assays shown in panel c, pre-loaded with either the 46 (TtCmr-46), 40 (TtCmr-40) or 34 nt (TtCmr-34) crRNA (top strand). Red triangles indicate the anticipated cleavage sites (α, β, γ and δ) in the 4.5 target RNA (bottom strand, Supplementary Table S1) by the endoribonuclease activity of the Cas7 subunits. The target RNA was radiolabeled at the 5′ end with 32P γ-ATP (“P” in the red circle). b Denaturing PAGE analysis of the activity assay using a 5′ labeled target RNA complementary to the crRNA incubated with the endogenous TtCmr complex. A single stranded RNA marker (“M”) was used as size standards as indicated on the left. c Activity assays similar to panel b but using the reconstituted complexes. Discontinuous gel lanes are indicated by a dashed line. The results of the cleavage assays are representative results of three (b) or two (c) replicates (Supplementary Fig. 6). Source data are provided as a Source data file.
