Fig. 1: Mapping 3D genome architecture through mouse spermatogenesis by Hi-C.
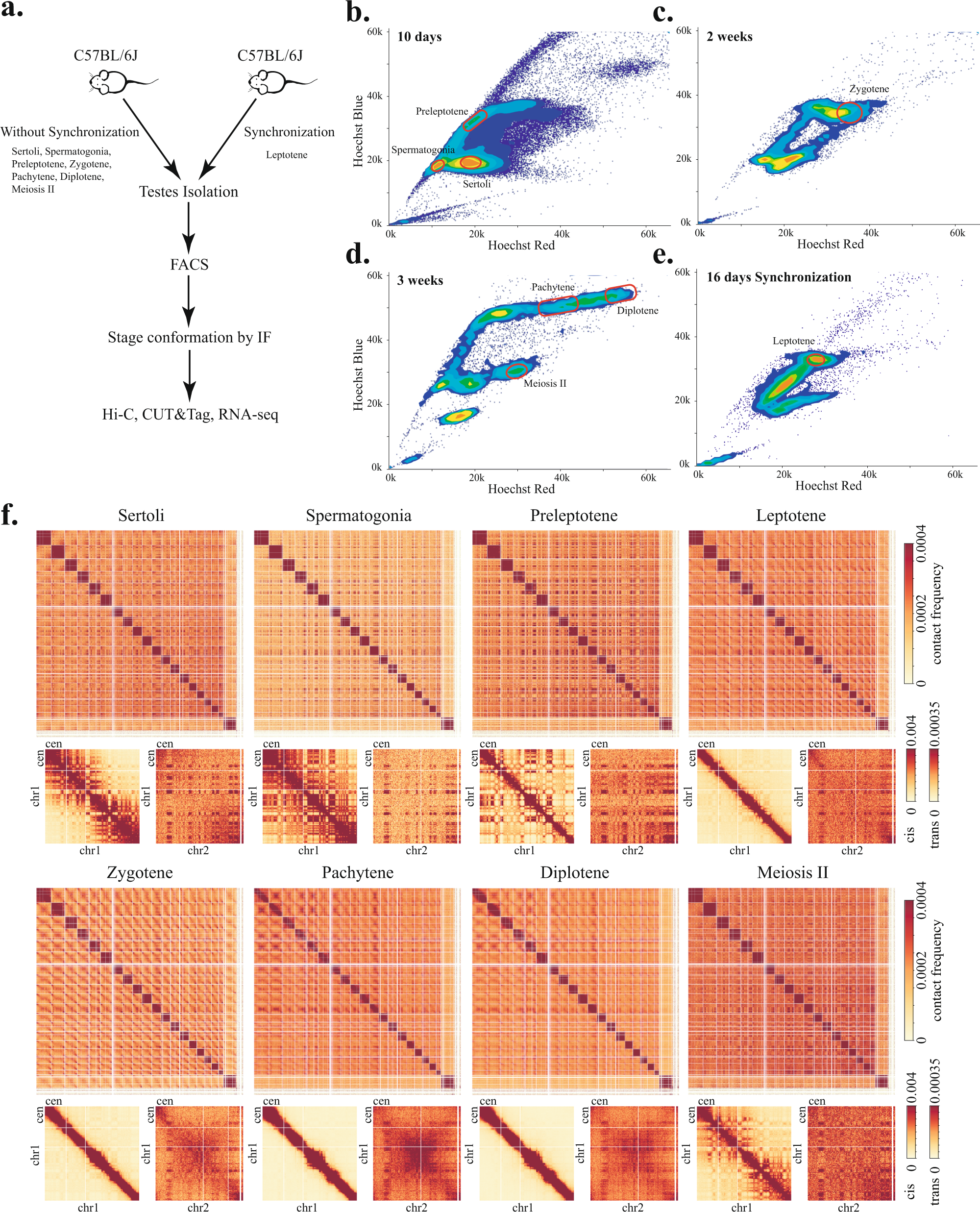
a Experimental workflow for isolating somatic cells and spermatocytes of different stages. b–e Representative Hoechst profiles show the separation of different cell types by fluorescence intensity in mice of different ages: 10-day-old mice for isolating Sertoli, spermatogonia, and preleptotene cells (b), 2-week-old mice for zygotene cells (c), 3-week-old mice for pachytene, diplotene, and meiosis II cells (d), 16 days synchronized mice for isolation of leptotene cells (e). Red circles in each profile indicate gating windows used for cell isolation. f Genome-wide Hi-C interaction heatmaps binned at 1 Mb resolution show dynamic reorganization of 3D genome architecture during meiosis. Enlarged Chr1 and Chr1–Chr2 trans interaction heatmaps at 1 Mb resolution are shown below the genome-wide heatmaps. Cen indicates the centromere ends of chromosomes.
