Fig. 1: Fully learned sequence and rotamer design onto fixed protein backbones.
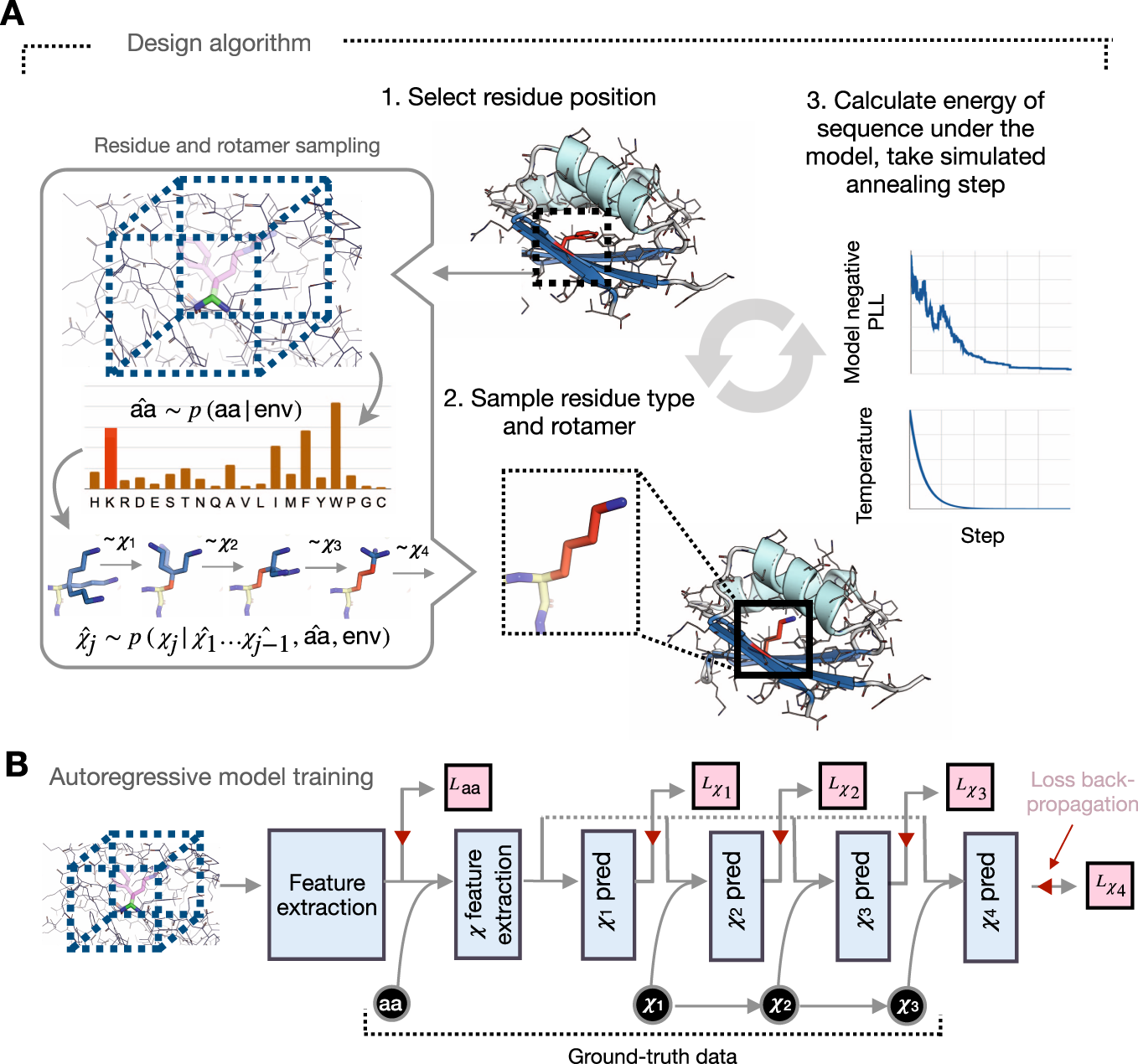
A Sequences are designed onto fixed protein backbones by (1) iteratively selecting a candidate residue position, (2) using a neural network model to sample amino-acid type and conformation, and (3) optimizing the negative pseudo-log-likelihood of the sequence under the model via simulated annealing. (Inset, left) Given the local chemical environment around a residue position (box, dashed, not to scale), residue type and rotamer angles are sampled from network-predicted distributions. B The neural network model is trained to predict residue identity and rotamer angles in an autoregressive fashion, conditioning on ground-truth data (black). The trained classifier predicts amino-acid type as well as rotamer angles conditioned on the amino-acid type. Cross-entropy loss objectives are shown in pink.
