Fig. 3: Pharmacological ER stress induction activates protein disaggregation machinery.
From: Stress-induced protein disaggregation in the endoplasmic reticulum catalysed by BiP
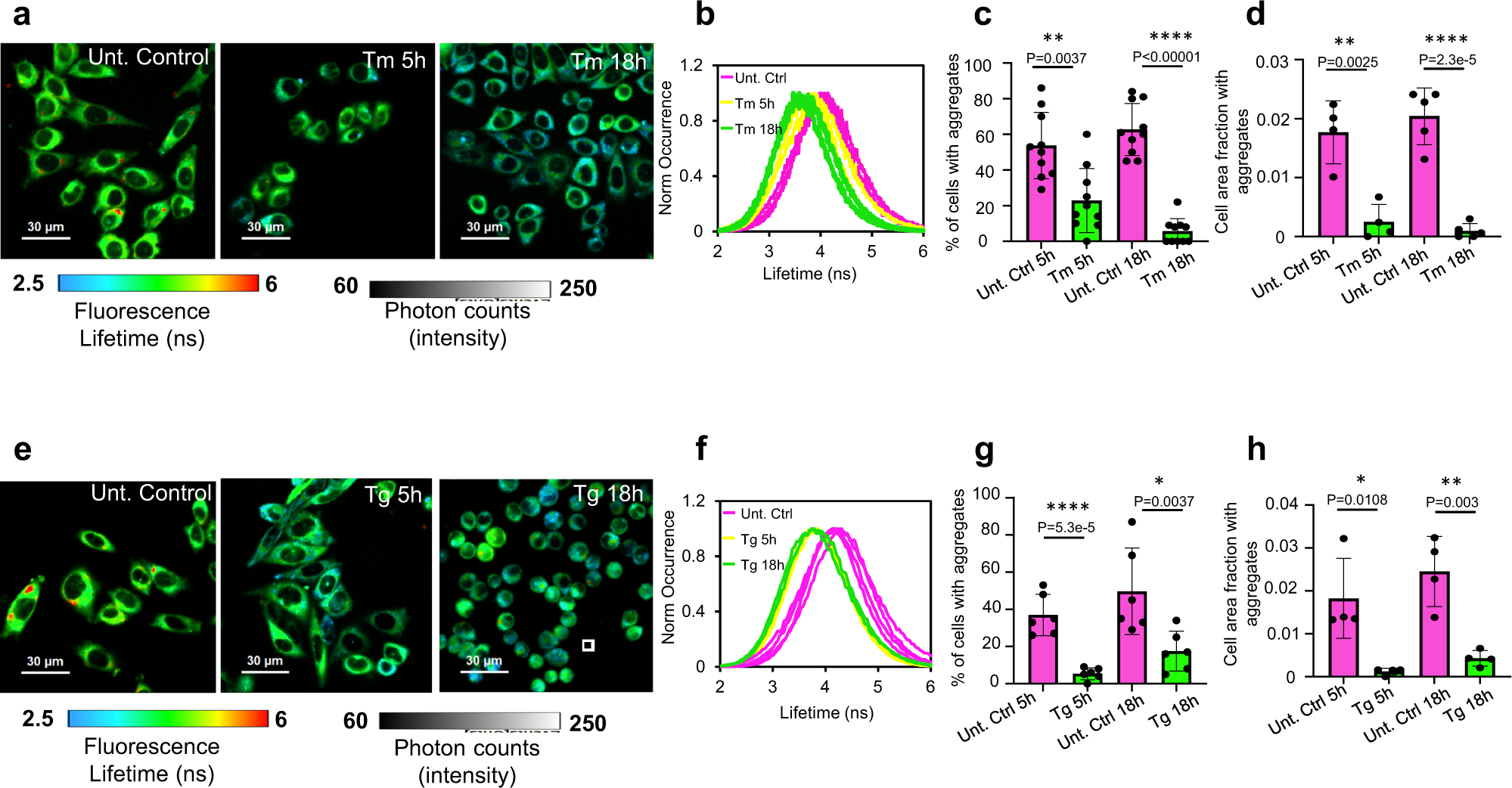
a, e FLIM images of live CHO-K1 cells stably expressing HT-aggrER, untreated or treated with tunicamycin (Tm at 0.5 µg/mL) or thapsigargin (Tg at 0.5 µM) for a time period post-pulse-labelling with the P1 fluorochrome as indicated. b, f Histograms representing frequency distribution of pixel fluorescence lifetime from image-series as in (a, e), note the shift towards shorter lifetime values post-treatment (traces represent individual cells, untreated control, pink; 5 h treatment, yellow; 18 h treatment, green). c, g Quantitation of the relative number of cells with FLIM-detected aggregates in images as in (a, e). d, h Plots of relative cell area occupied by FLIM-detected aggregates (lifetime > 5 ns, red puncta) in images as in (a, e). Mean ± SEM, n = 10, 5, 6, 4 independent fields of view containing at least 12 cells each (c, d, g, h), respectively. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001, unpaired T test (two-tailed).
