Fig. 1: Between database comparisons with ATLAS.
From: Seeking patterns of antibiotic resistance in ATLAS, an open, raw MIC database with patient metadata
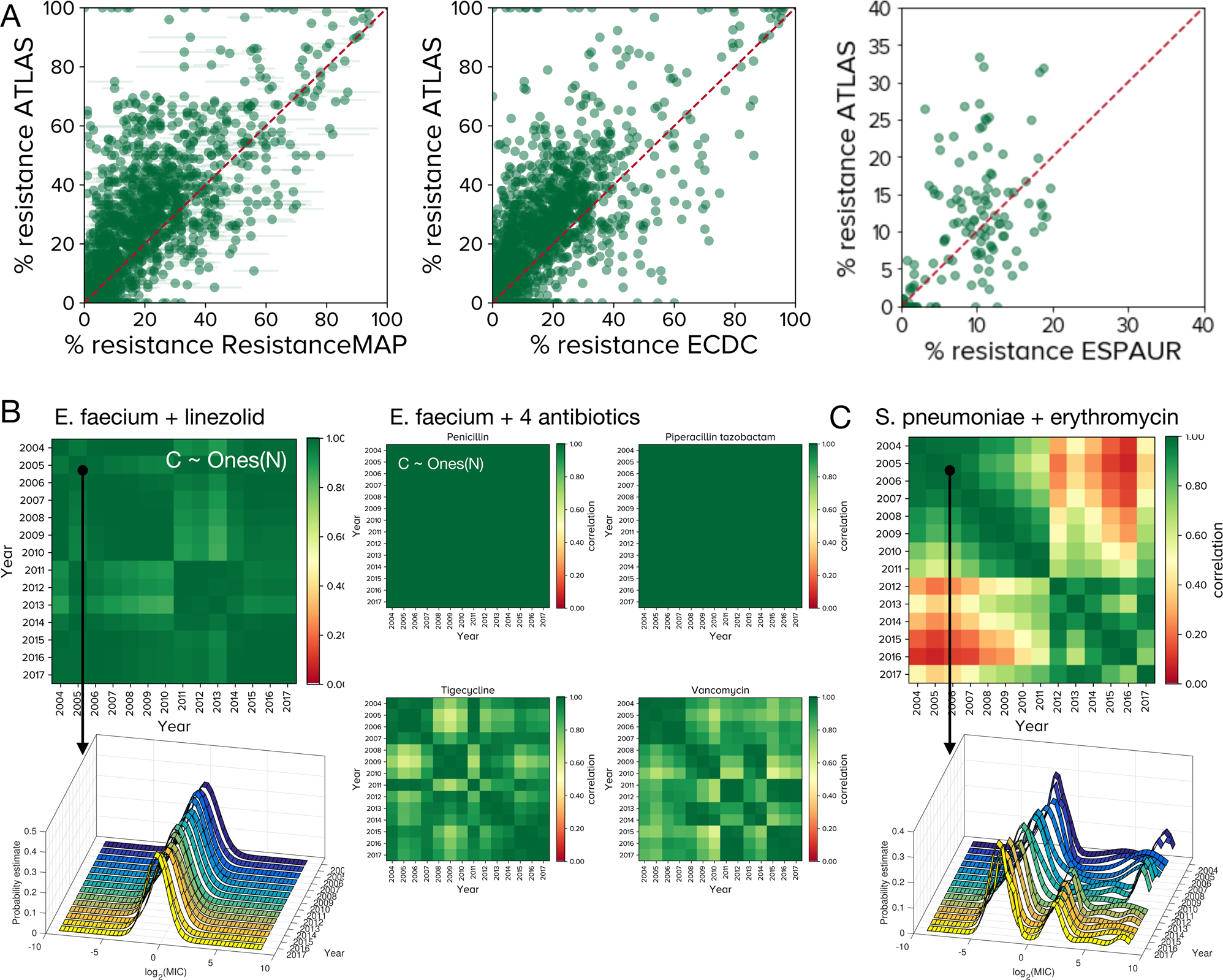
A Each point represents a PA (pathogen-antibiotic) pair in a given country, for a given year with frequency of resistance (fR as a %age) on x and y axes. ATLAS tends to over-estimates fR relative to ResistanceMap, ECDC and ESPAUR data: differences between fR in ATLAS and other databases are positively skewed, however between-database differences are smaller for larger PA pair datasets (Supplementary Fig. 2 has statistics). B Between-year correlations for many ATLAS PA pairs form correlelograms, called `C' here, that are close to the (pure green) unity matrix of ones, Ones(N), for N years. 5 PA pairs for Enterococcus faecium are shown. (Supplementary Fig. 8C has some correlelogram statistics, Supplementary Fig. 9 shows many are close to Ones(N) but not all, see Fig. 3 and Supplementary Fig. 10). The left column shows the global MIC (minimal inhibitory concentration) distribution of E. faecium and linezolid is stable from year to year and its correlelogram is close to Ones(N). The middle panel shows 4 correlelograms with banded structures that occur when MIC distributions experience change. C This correlelogram of Streptococcus pneumoniae and erythromycin have a block structure because their MIC distributions correlate poorly between years: a high-MIC cluster diminishes and is replaced by a cluster with lower MIC in 2010–2011 (c.f. Fig. 4).
