Fig. 1: Laminar shear stress increases chromatin accessibility and vasculo-protective gene expression.
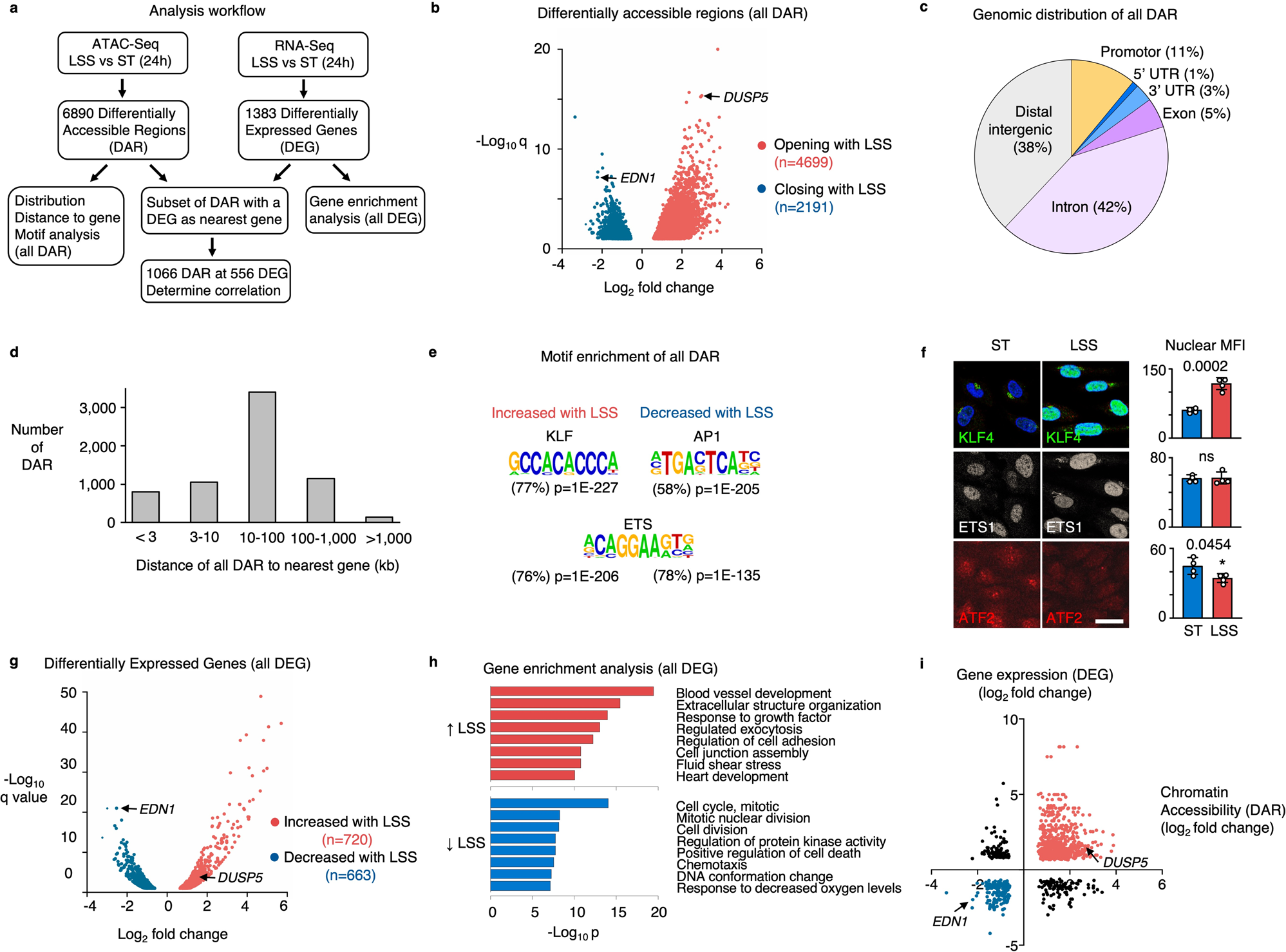
a Schematic illustration of the workflow and analysis. b Volcano plot showing the differentially accessible regions (DAR) determined by ATAC-Seq in PAEC that were exposed to 15 dyn/cm2 of LSS for 24 h vs static controls. Each dot represents a DAR. Red dots are DAR with increased accessibility, blue dots are DAR with decreased accessibility under LSS. Highlighted genes are described in subsequent sections. n = 3 experimental replicates. P values were determined by a two-tailed Wald test with Benjamini–Hochberg adjustment. c Pie chart showing the annotation of DAR peaks, analyzed using Homer. d Bar graphs showing the distance of DAR to its nearest gene, analyzed using Homer. e Motif enrichment of DAR with increased or decreased accessibility with LSS, analyzed using Homer. P values were determined by binomial test. f Representative immunofluorescent images (left) with quantitation on the right, of PAEC that were exposed to 15 dyn/cm2 of LSS for 24 h vs static controls (ST), showing the nuclear expression of KLF4 (green); ETS1 (gray), and ATF2 (red). Nuclei were stained with DAPI, (blue). n = 4 experimental replicates. Bars indicate mean ± s.e.m. P values were determined by Student’s two-tailed t-test. ns: not significant. Source data are provided as a Source Data file. Scale bar, 20 μm. g Volcano plot showing the differentially expressed genes (DEG) determined by RNA-Seq in PAEC exposed to 15 dyn/cm2 of LSS for 24 h vs Static controls. Each dot represents a gene. Red dots are DEG with increased expression under LSS, blue dots are DEG with decreased expression under LSS. Highlighted genes are described in subsequent sections. n = 2 experimental replicates. P values were determined by a two-tailed Wald test with Benjamini–Hochberg adjustment. h Gene enrichment analysis using Metascape of genes that are induced by LSS (red bars) and genes that are repressed by LSS (blue bars). P values were determined by a one-tailed hypergeometric test with Benjamin-Hochberg adjustment. i Scatterplot showing a positive correlation of accessibility changes with the expression of its nearest differentially expressed gene. Highlighted genes are described in subsequent sections. r = 0.41 with P < 0.0001 determined by two-tailed Pearson R test.
