Fig. 1: TcdB subtypes show variable dependency on FZD and CSPG4 receptors.
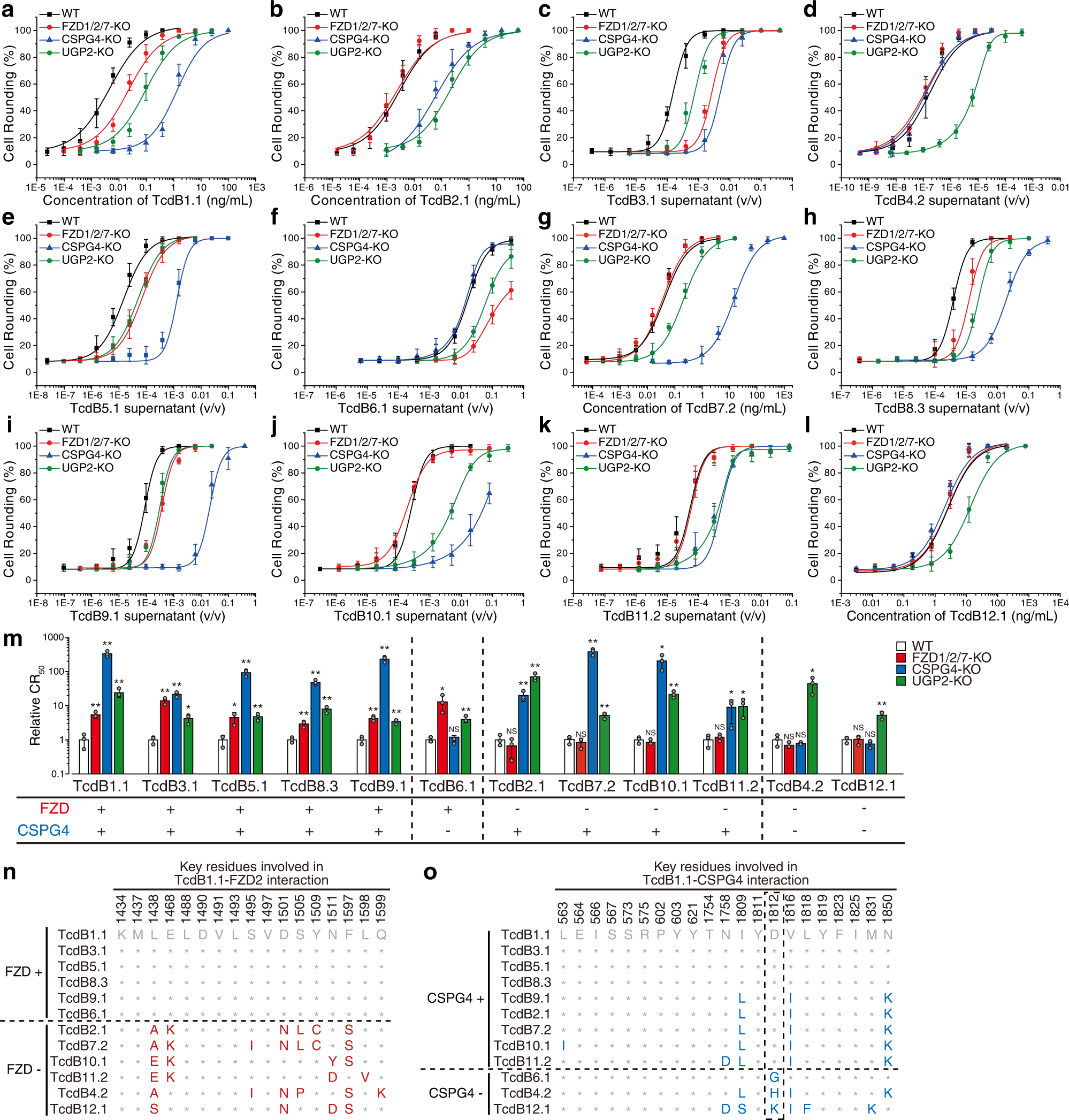
HeLa wildtype (WT), stable FZD1/2/7 knockout (FZD1/2/7-KO), CSPG4-KO, and UGP2-KO cells were exposed to either recombinant TcdB1.1 (a), TcdB2.1 (b), TcdB7.2 (g), and TcdB12.1 (l), or culture supernatants from native C. difficile strains expressing TcdB3.1 (c), TcdB4.2 (d), TcdB5.1 (e), TcdB6.1 (f), TcdB8.3 (h), TcdB9.1 (i), TcdB10.1 (j), and TcdB11.2 (k) for 24 h. The percentages of round-shaped cells were plotted over toxin concentrations or supernatant dilutions. Error bars indicate mean ± s.d., N = 3 (biologically independent experiments). Strain information is listed in Supplementary Table 1. The activity of TcdB in each supernatant has been validated using a polyclonal TcdB antibody as shown in Supplementary Fig. 1. m Summary of the receptor preference of TcdB variants. The toxin concentrations inducing 50% of cell rounding (CR50) were determined. The relative CR50 values in different cell lines were normalized to WT and plotted as a bar-chart. The dependency on FZDs and CSPG4 are noted with plus and minus signs. Error bars indicate mean ± s.d.; N = 3 (biologically independent experiments); *, p < 0.05; **, p < 0.01; NS not significant (Student’s t-test, two-sided). n A list of residues across tested TcdB subtypes at 17 key positions mediating TcdB1.1-FZD2 interactions. These positions are based on the crystal structure of TcdB-FZD complex (PDB: 6C0B)27. o A list of residues across tested TcdB subtypes at 21 key positions mediating TcdB1.1-CSPG4 interactions. These positions are based on the cryo-EM structure of TcdB-CSPG4 complex (PDB: 7ML7)28. Residue 1812 was highlighted with a dash box. Source data are provided as a Source Data file.
