Fig. 1: Fast imaging of live cell anatomy of 15 intracellular structures by the universal lipid staining and deep learning segmentation using a spinning disk confocal microscope with an extended resolution.
From: Fast segmentation and multiplexing imaging of organelles in live cells
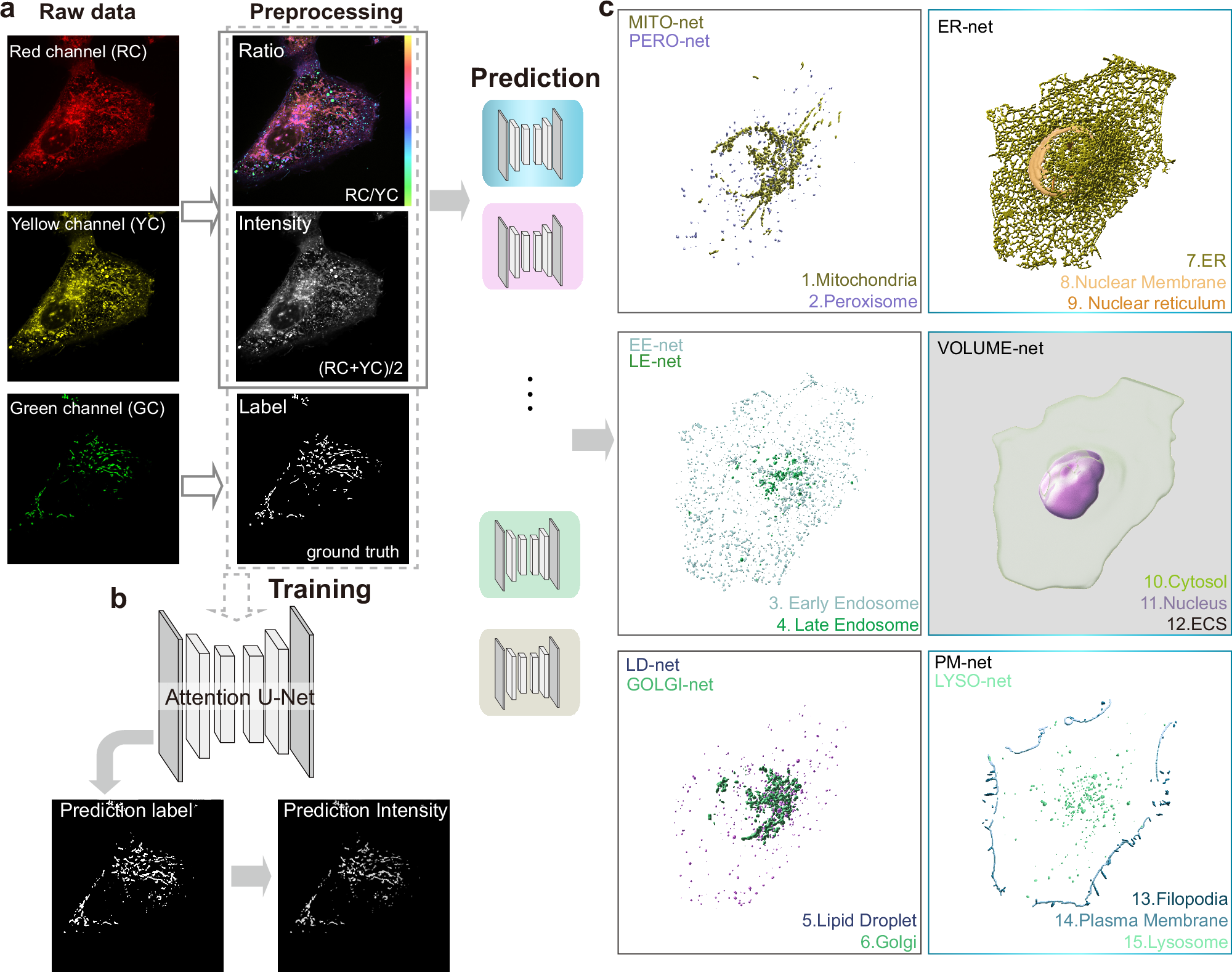
a Schematic diagram of data acquisition and preprocessing. Extended-resolution image of U2-OS cells labeled by both the organelle-specific GFP and Nile Red are imaged with the same excitation 488 nm and three detection channels (green, yellow, and red). The spectral ratio and intensity are calculated by the yellow channel (Em: 580–653 nm) and the red channel (Em: 665–705 nm). The ground truth label is the binary mask of green channel (Em: 500–550 nm). b The untrained Attention U-Net was fed with the ratio, intensity, and label image to train neural networks for distinct organelles. Upon completion of the training process, the model will output the prediction label which will multiply the intensity image to get the prediction intensity. c From the super-resolution intensity image and the spectral ratio image, the deep convolutional neural networks (DCNN) predict binary masks of 15 subcellular structures. The masks of cytosol, nucleus, and extracellular space are segmented by VOLUME-net. The masks of ER, nuclear reticulum, and nuclear membrane are segmented by ER-net. The Golgi mask is segmented by GOLGI-net and the lipid droplet mask is segmented by LD-net. Mitochondria mask segmented by MITO-net and lysosome mask segmented by LYSO-net. Early endosome mask segmented by EE-net and late endosome mask segmented by LE-net. Peroxisome mask segmented by PERO-net and the masks of the plasma membrane and filopodia segmented by PM-net (Supplementary Table 1).
