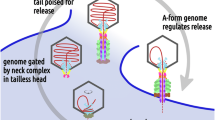
Abstract
The post-genome packaging mechanisms that govern the assembly of an infectious virion are poorly understood in bacteriophages and other viruses. Here, our near-atomic resolution cryo-EM structural analyses uncovered an assembly- and conformation-driven genome positioning mechanism in the tailed bacteriophage T4. We show that following headful packaging, which generates a pressurized head, a global conformational change occurs in the portal structure, probably triggering packaging termination and ejection of the packaging motor. Our high-resolution structures of the neck of the virion further show that the neck undergoes conformational changes upon docking of a pre-assembled tail onto the sealed neck, which then opens a genome-gate. Driven by the pressure of the packaged DNA, the genome travels through open neck channels, binds and compresses the resident tape-measure protein, and halts at the bottom of the second topmost disk of the tail tube. Pressure-suspended within the virion’s innermost tunnel and secured by a baseplate plug, the genome remains poised to flow through the tunnel into a host cell upon receiving the host receptor recognition signal.
Similar content being viewed by others
Data availability
The C1 and C6 symmetric cryo-EM reconstructions focused on the phage neck region have been deposited in the EM Data Bank with the accession codes EMD-48458 and EMD-48462, respectively. The C3 and C6 symmetric reconstructions of the middle part of the tail have been deposited with the accession codes EMD-48460 and EMD-48463, respectively; and the C3 and C6 symmetric reconstructions of the distal part of the tail have been deposited with the accession codes EMD-48459 and EMD-48464, respectively. The composite map of the whole portal-neck-tail complex has been deposited with the accession code EMD-48324. The asymmetric atomic structure of the neck has been deposited in the Protein Data Bank (PDB) with the accession code 9MOF. The three-fold-symmetric structures of the middle and distal parts of the tail have been deposited with the PDB accession codes 9MOH and 9MOG, respectively. The composite structure of the whole portal-neck-tail complex has been deposited with the PDB accession code 9MKB.
References
Earnshaw, W. C. & Casjens, S. R. DNA packaging by the double-stranded DNA bacteriophages. Cell 21, 319–331 (1980).
Han, L. et al. Cryo-EM structures of bacteriophage T4 portal-neck assembly intermediates reveal a viral genome retention mechanism. Nat. Commun. https://doi.org/10.1038/s41467-026-69107-7 (2026). Epub ahead of print.
Leiman, P. G., Kanamaru, S., Mesyanzhinov, V. V., Arisaka, F. & Rossmann, M. G. Structure and morphogenesis of bacteriophage T4. Cell Mol. Life Sci. 60, 2356–2370 (2003).
Leiman, P. G. et al. Morphogenesis of the T4 tail and tail fibers. Virol. J. 7, 355 (2010).
Leiman, P. G. & Shneider, M. M. Contractile tail machines of bacteriophages. Adv. Exp. Med. Biol. 726, 93–114 (2012).
Fokine, A. & Rossmann, M. G. Molecular architecture of tailed double-stranded DNA phages. Bacteriophage 4, e28281 (2014).
Leiman, P. G., Chipman, P. R., Kostyuchenko, V. A., Mesyanzhinov, V. V. & Rossmann, M. G. Three-dimensional rearrangement of proteins in the tail of bacteriophage T4 on infection of its host. Cell 118, 419–429 (2004).
Taylor, N. M. et al. Structure of the T4 baseplate and its function in triggering sheath contraction. Nature 533, 346–352 (2016).
Yap, M. L. et al. Role of bacteriophage T4 baseplate in regulating assembly and infection. Proc. Natl. Acad. Sci. USA. 113, 2654–2659 (2016).
Fang, Q. et al. Structural morphing in a symmetry-mismatched viral vertex. Nat. Commun. 11, 1713 (2020).
Rao, V. B., Fokine, A., Fang, Q. & Shao, Q. Bacteriophage T4 Head: Structure, Assembly, and Genome Packaging. Viruses 15, 10.3390/v15020527 (2023).
Black, L. W. & Rao, V. B. Structure, assembly, and DNA packaging of the bacteriophage T4 head. Adv. Virus Res. 82, 119–153 (2012).
Rao, V. B. & Feiss, M. Mechanisms of DNA packaging by large double-stranded DNA viruses. Annu Rev. Virol. 2, 351–378 (2015).
Black, L. W. & Thomas, J. A. Condensed genome structure. Adv. Exp. Med Biol. 726, 469–487 (2012).
Liu, S. et al. A viral packaging motor varies its DNA rotation and step size to preserve subunit coordination as the capsid fills. Cell 157, 702–713 (2014).
Berndsen, Z. T., Keller, N. & Smith, D. E. Continuous allosteric regulation of a viral packaging motor by a sensor that detects the density and conformation of packaged DNA. Biophys. J. 108, 315–324 (2015).
Villanueva Valencia, J. R., Li, D., Casjens, S. R. & Evilevitch, A. SAXS-osmometer’ method provides measurement of DNA pressure in viral capsids and delivers an empirical equation of state. Nucleic Acids Res. 51, 11415–11427 (2023).
Sun, L. et al. Cryo-EM structure of the bacteriophage T4 portal protein assembly at near-atomic resolution. Nat. Commun. 6, 7548 (2015).
Rao, V. B., Fokine, A. & Fang, Q. The remarkable viral portal vertex: structure and a plausible model for mechanism. Curr. Opin. Virol. 51, 65–73 (2021).
Padilla-Sanchez, V. et al. Structure-function analysis of the DNA translocating portal of the bacteriophage T4 packaging machine. J. Mol. Biol. 426, 1019–1038 (2014).
Grimes, S., Ma, S., Gao, J., Atz, R. & Jardine, P. J. Role of φ29 connector channel loops in late-stage DNA packaging. J. Mol. Biol. 410, 50–59 (2011).
Zhang, Z. et al. A promiscuous DNA packaging machine from bacteriophage T4. PLoS Biol. 9, e1000592 (2011).
Prevo, B. & Earnshaw, W. C. DNA packaging by molecular motors: from bacteriophage to human chromosomes. Nat. Rev. Genet. 25, 785–802 (2024).
Ivanovska, I., Wuite, G., Jönsson, B. & Evilevitch, A. Internal DNA pressure modifies stability of WT phage. Proc. Natl. Acad. Sci. USA. 104, 9603–9608 (2007).
Lhuillier, S. et al. Structure of bacteriophage SPP1 head-to-tail connection reveals mechanism for viral DNA gating. Proc. Natl. Acad. Sci. USA. 106, 8507–8512 (2009).
Orlov, I. et al. CryoEM structure and assembly mechanism of a bacterial virus genome gatekeeper. Nat. Commun. 13, 7283 (2022).
Cardarelli, L. et al. The crystal structure of bacteriophage HK97 gp6: defining a large family of head-tail connector proteins. J. Mol. Biol. 395, 754–768 (2010).
Wang, Z. et al. Structure of vibrio phage XM1, a simple contractile DNA injection machine. Viruses 15, 10.3390/v15081673 (2023).
Olia, A. S., Prevelige, P. E., Johnson, J. E. & Cingolani, G. Three-dimensional structure of a viral genome-delivery portal vertex. Nat. Struct. Mol. Biol. 18, 597–603 (2011).
Fokine, A. et al. The molecular architecture of the bacteriophage T4 neck. J. Mol. Biol. 425, 1731–1744 (2013).
Akhter, T. et al. The neck of bacteriophage T4 is a ring-like structure formed by a hetero-oligomer of gp13 and gp14. Biochim Biophys. Acta 1774, 1036–1043 (2007).
Aksyuk, A. A. et al. The tail sheath structure of bacteriophage T4: A molecular machine for infecting bacteria. EMBO J. 28, 821–829 (2009).
Taylor, N. M. I., van Raaij, M. J. & Leiman, P. G. Contractile injection systems of bacteriophages and related systems. Mol. Microbiol 108, 6–15 (2018).
Zhao, L., Kanamaru, S., Chaidirek, C. & Arisaka, F. P15 and P3, the tail completion proteins of bacteriophage T4, both form hexameric rings. J. Bacteriol. 185, 1693–1700 (2003).
Sonani, R. R. et al. Neck and capsid architecture of the robust agrobacterium phage milano. Commun. Biol. 6, 921 (2023).
Li, F. et al. High-resolution cryo-EM structure of the pseudomonas bacteriophage E217. Nat. Commun. 14, 4052 (2023).
Yang, F., Wang, L., Zhou, J., Xiao, H. & Liu, H. In situ structures of the ultra-long extended and contracted tail of myoviridae phage P1.Viruses 15, 10.3390/v15061267 (2023).
Abuladze, N. K., Gingery, M., Tsai, J. & Eiserling, F. A. Tail length determination in bacteriophage T4. Virology 199, 301–310 (1994).
Duda, R. L., Gingery, M., Ishimoto, L. K. & Eiserling, F. A. Expression of plasmid-encoded structural proteins permits engineering of bacteriophage T4 assembly. Virology 179, 728–737 (1990).
Arisaka, F., Yap, M. L., Kanamaru, S. & Rossmann, M. G. Molecular assembly and structure of the bacteriophage T4 tail. Biophys. Rev. 8, 385–396 (2016).
Mahony, J. et al. Functional and structural dissection of the tape measure protein of lactococcal phage TP901-1. Sci. Rep. 6, 36667 (2016).
Katsura, I. & Hendrix, R. W. Length determination in bacteriophage lambda tails. Cell 39, 691–698 (1984).
Kizziah, J. L., Mukherjee, A., Parker, L. K. & Dokland, T. Structure of the Staphylococcus aureus bacteriophage 80α neck shows details of the DNA, tail completion protein, and tape measure protein. Structure 33, 1063–1073 (2025).
Xiao, H. et al. Structure of the siphophage neck-Tail complex suggests that conserved tail tip proteins facilitate receptor binding and tail assembly. PLoS Biol. 21, e3002441 (2023).
Linares, R. & Breyton, C. About bacteriophage tail terminator and tail completion proteins: structure of the proximal extremity of siphophage T5 tail. J. Virol. 99, e0137624 (2025).
Shao, Q. et al. Cryo-EM structures of phage T4 infection intermediate uncover a genome piloting mechanism. Submitted.
Duda, R. L., Wall, J. S., Hainfeld, J. F., Sweet, R. M. & Eiserling, F. A. Mass distribution of a probable tail-length-determining protein in bacteriophage T4. Proc. Natl. Acad. Sci. USA. 82, 5550–5554 (1985).
Rao, V. B. A virus DNA gate: zipping and unzipping the packed viral genome. Proc. Natl. Acad. Sci. USA. 106, 8403–8404 (2009).
Oliveira, L., Tavares, P. & Alonso, J. C. Headful DNA packaging: bacteriophage SPP1 as a model system. Virus Res. 173, 247–259 (2013).
Bayfield, O. W., Steven, A. C. & Antson, A. A. Cryo-EM structure in situ reveals a molecular switch that safeguards virus against genome loss. eLife 9, e55517 (2020).
Hu, B., Margolin, W., Molineux, I. J. & Liu, J. Structural remodeling of bacteriophage T4 and host membranes during infection initiation. Proc. Natl. Acad. Sci. USA. 112, E4919–E4928 (2015).
Tao, P., Mahalingam, M. & Rao, V. B. Highly effective soluble and bacteriophage t4 nanoparticle plague vaccines against yersinia pestis. Methods Mol. Biol. 1403, 499–518 (2016).
Tao, P., Li, Q., Shivachandra, S. B. & Rao, V. B. Bacteriophage T4 as a nanoparticle platform to display and deliver pathogen antigens: Construction of an effective anthrax vaccine. Methods Mol. Biol. 1581, 255–267 (2017).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Rohou, A. & Grigorieff, N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Kimanius, D., Dong, L., Sharov, G., Nakane, T. & Scheres, S. H. W. New tools for automated cryo-EM single-particle analysis in RELION-4.0. Biochem J. 478, 4169–4185 (2021).
Fokine, A. et al. Molecular architecture of the prolate head of bacteriophage T4. Proc. Natl. Acad. Sci. USA. 101, 6003–6008 (2004).
Henderson, R. et al. Outcome of the first electron microscopy validation task force meeting. Structure 20, 205–214 (2012).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of coot. Acta Crystallogr D. Biol. Crystallogr 66, 486–501 (2010).
Boudko, S. P., Strelkov, S. V., Engel, J. & Stetefeld, J. Design and crystal structure of bacteriophage T4 mini-fibritin NCCF. J. Mol. Biol. 339, 927–935 (2004).
Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr D. Struct. Biol. 74, 531–544 (2018).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr D. Struct. Biol. 75, 861–877 (2019).
Goddard, T. D. et al. UCSF chimerax: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018).
Pettersen, E. F. et al. UCSF chimerax: Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Acknowledgements
This work was supported by NIAID, NIH grant 1R01AI175340 to V.B.R. and A.F. and in part by NIDA, NIH Avant Garde Award 1DP1DA060580 and National Science Foundation grant MCB-0923873 to V.B.R. Research in Q.F.’s laboratory was supported by the National Natural Science Foundation of China (32371285).
Author information
Authors and Affiliations
Contributions
V.B.R., M.G.R., and A.F. conceived the project. A.F., J.Z., T.K., F.V., C-A.A., Z.W., B.K., and V.B.R. performed research. A.F., J.Z., C-A.A., M.G.R., Z.C., L.S., Q.F., R.J.K. and V.B.R. analyzed data. V.B.R., R.J.K., and M.G.R. supervised the project. A.F. and V.B.R. prepared the manuscript, with additional edits from all authors. V.B.R. provided overall direction and coordination for this and the accompanying genome retention project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Alfred Antson who co-reviewed with Brianna Woodbury, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Fokine, A., Zhu, J., Klose, T. et al. In situ structures of the portal-neck-tail complex of bacteriophage T4 inform a viral genome positioning mechanism. Nat Commun (2026). https://doi.org/10.1038/s41467-026-69106-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-026-69106-8