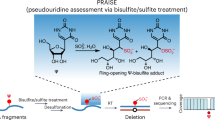
Abstract
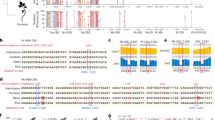
Pseudouridine (Ψ) is an abundant modification in small RNA catalyzed by multiple pseudouridine synthases (PUSs). However, the substrate specificity of human PUSs remains elusive. Here, we adopted PRAISE, a quantitative Ψ detection method, to profile pseudouridylation in small RNA, including cytosolic and mitochondrial tRNAs, snRNA, and snoRNA. We found that snoRNA pseudouridylation is mediated not only by RNA-guided DKC1, but also by the stand-alone enzyme PUS7 at a specific site. Interestingly, several PUS enzymes, including PUS1, RPUSD1, and PUS7, which install nearby Ψ sites within tRNA anticodon stem-loop, can influence pseudouridylation catalyzed by other PUSs, revealing an unrecognized interplay during Ψ formation. For the three RluA family enzymes, RPUSD1 catalyzes the canonical Ψ30 in tRNA-Ile and Ψ72 in tRNA-Arg isoacceptors. RPUSD2 pseudouridylates Ψ31 of mt-tRNALeu(CUN), Ψ32 of mt-tRNAPro and mt-tRNACys, whereas RPUSD3 lacks tRNA activity. Together, our quantitative Ψ profiling characterized PUS tRNA substrates and revealed unexpected PUS interplay.
Similar content being viewed by others
Data availability
The sequencing data supporting the results of this study are available in public repositories. The main raw and processed sequencing data have been deposited into the NCBI Gene Expression Omnibus (GEO) under the accession number GSE299274. Additional sequencing data have been deposited in the Genome Sequence Archive (GSA) at the CNCB-NGDC under PRJCA050602. Source data are provided with this paper.
Code availability
The analysis scripts in this study are publicly available in the GitHub repository at https://github.com/LiuWenQing657/PsiUInterplayAnalysis, which is also cited here71.
References
Li, X., Ma, S. & Yi, C. Pseudouridine: the fifth RNA nucleotide with renewed interests. Curr. Opin. Chem. Biol. 33, 108–116 (2016).
Borchardt, E. K., Martinez, N. M. & Gilbert, W. V. Regulation and function of RNA pseudouridylation in human cells. Annu. Rev. Genet. 54, 309–336 (2020).
Charette, M. & Gray, M. W. Pseudouridine in RNA: what, where, how, and why. IUBMB Life 49, 341–351 (2000).
Luo, N., Huang, Q., Zhang, M. & Yi, C. Functions and therapeutic applications of pseudouridylation. Nat. Rev. Mol. Cell Biol. 26, 691–705 (2025).
Cerneckis, J., Cui, Q., He, C., Yi, C. & Shi, Y. Decoding pseudouridine: an emerging target for therapeutic development. Trends Pharmacol. Sci. 43, 522–535 (2022).
Ge, J. & Yu, Y. T. RNA pseudouridylation: new insights into an old modification. Trends Biochem. Sci. 38, 210–218 (2013).
Suzuki, T. The expanding world of tRNA modifications and their disease relevance. Nat. Rev. Mol. Cell Biol. 22, 375–392 (2021).
Spenkuch, F., Motorin, Y. & Helm, M. Pseudouridine: still mysterious, but never a fake (uridine)!. RNA Biol. 11, 1540–1554 (2014).
Patton, J. R., Bykhovskaya, Y., Mengesha, E., Bertolotto, C. & Fischel-Ghodsian, N. Mitochondrial myopathy and sideroblastic anemia (MLASA): missense mutation in the pseudouridine synthase 1 (PUS1) gene is associated with the loss of tRNA pseudouridylation. J. Biol. Chem. 280, 19823–19828 (2005).
Fernandez-Vizarra, E., Berardinelli, A., Valente, L., Tiranti, V. & Zeviani, M. Nonsense mutation in pseudouridylate synthase 1 (PUS1) in two brothers affected by myopathy, lactic acidosis and sideroblastic anaemia (MLASA). J. Med. Genet. 44, 173–180 (2007).
Bykhovskaya, Y., Casas, K., Mengesha, E., Inbal, A. & Fischel-Ghodsian, N. Missense mutation in pseudouridine synthase 1 (PUS1) causes mitochondrial myopathy and sideroblastic anemia (MLASA). Am. J. Hum. Genet. 74, 1303–1308 (2004).
Wang, B. et al. Mitochondrial tRNA pseudouridylation governs erythropoiesis. Blood 144, 657–671 (2024).
Shaheen, R. et al. PUS7 mutations impair pseudouridylation in humans and cause intellectual disability and microcephaly. Hum. Genet. 138, 231–239 (2019).
Guzzi, N. et al. Pseudouridylation of tRNA-derived fragments steers translational control in stem cells. Cell 173, 1204–1216 e1226 (2018).
Cui, Q. et al. Targeting PUS7 suppresses tRNA pseudouridylation and glioblastoma tumorigenesis. Nat. Cancer 2, 932–949 (2021).
Bakin, A. & Ofengand, J. Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyltransferase center: analysis by the application of a new sequencing technique. Biochemistry 32, 9754–9762 (1993).
Li, X. et al. Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat. Chem. Biol. 11, 592–597 (2015).
Lovejoy, A. F., Riordan, D. P. & Brown, P. O. Transcriptome-wide mapping of pseudouridines: pseudouridine synthases modify specific mRNAs in S. cerevisiae. PLoS ONE 9, e110799 (2014).
Schwartz, S. et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 159, 148–162 (2014).
Carlile, T. M. et al. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 515, 143–146 (2014).
Song, J. et al. Differential roles of human PUS10 in miRNA processing and tRNA pseudouridylation. Nat. Chem. Biol. 16, 160–169 (2020).
Lucas, M. C. et al. Quantitative analysis of tRNA abundance and modifications by nanopore RNA sequencing. Nat. Biotechnol. 42, 72–86 (2024).
Zhang, M. et al. Quantitative profiling of pseudouridylation landscape in the human transcriptome. Nat. Chem. Biol. 19, 1185–1195 (2023).
Dai, Q. et al. Quantitative sequencing using BID-seq uncovers abundant pseudouridines in mammalian mRNA at base resolution. Nat. Biotechnol. 41, 344–354 (2023).
Xu, H. et al. Absolute quantitative and base-resolution sequencing reveals comprehensive landscape of pseudouridine across the human transcriptome. Nat. Methods 21, 2024–2033 (2024).
Zheng, G. et al. Efficient and quantitative high-throughput tRNA sequencing. Nat. Methods 12, 835–837 (2015).
Suzuki, T. et al. Complete chemical structures of human mitochondrial tRNAs. Nat. Commun. 11, 4269 (2020).
Sprinzl, M., Horn, C., Brown, M., Ioudovitch, A. & Steinberg, S. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 26, 148–153 (1998).
Blanchet, S. et al. Deciphering the reading of the genetic code by near-cognate tRNA. Proc. Natl. Acad. Sci. USA 115, 3018–3023 (2018).
Morais, P., Adachi, H. & Yu, Y. T. Spliceosomal snRNA Epitranscriptomics. Front. Genet. 12, 652129 (2021).
Ganot, P., Bortolin, M. L. & Kiss, T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell 89, 799–809 (1997).
Ni, J., Tien, A. L. & Fournier, M. J. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell 89, 565–573 (1997).
Karijolich, J. & Yu, Y. T. Spliceosomal snRNA modifications and their function. RNA Biol. 7, 192–204 (2010).
Nagpal, N., Tai, A. K., Nandakumar, J. & Agarwal, S. Domain specific mutations in dyskerin disrupt 3’ end processing of scaRNA13. Nucleic Acids Res. 50, 9413–9425 (2022).
Jorjani, H. et al. An updated human snoRNAome. Nucleic Acids Res. 44, 5068–5082 (2016).
Lestrade, L. & Weber, M. J. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res. 34, D158–D162 (2006).
Wu, G., Xiao, M., Yang, C. & Yu, Y. T. U2 snRNA is inducibly pseudouridylated at novel sites by Pus7p and snR81 RNP. EMBO J. 30, 79–89 (2011).
Chen, J. et al. m(6)A Regulates neurogenesis and neuronal development by modulating histone methyltransferase Ezh2. Genomics Proteomics Bioinformatics 17, 154–168 (2019).
Maldonado Lopez, A. M. et al. Mettl3-catalyzed m(6)A regulates histone modifier and modification expression in self-renewing somatic tissue. Sci. Adv. 9, eadg5234 (2023).
Behm-Ansmant, I., Grosjean, H., Massenet, S., Motorin, Y. & Branlant, C. Pseudouridylation at position 32 of mitochondrial and cytoplasmic tRNAs requires two distinct enzymes in Saccharomyces cerevisiae. J. Biol. Chem. 279, 52998–53006 (2004).
Levi, O. & Arava, Y. S. Pseudouridine-mediated translation control of mRNA by methionine aminoacyl tRNA synthetase. Nucleic Acids Res. 49, 432–443 (2021).
Zaganelli, S. et al. The pseudouridine synthase RPUSD4 is an essential component of mitochondrial RNA granules. J. Biol. Chem. 292, 4519–4532 (2017).
Fukasawa, Y. et al. MitoFates: improved prediction of mitochondrial targeting sequences and their cleavage sites. Mol. Cell Proteomics 14, 1113–1126 (2015).
Behm-Ansmant, I. et al. The Saccharomyces cerevisiae U2 snRNA:pseudouridine-synthase Pus7p is a novel multisite-multisubstrate RNA:Psi-synthase also acting on tRNAs. RNA 9, 1371–1382 (2003).
Lin, T. Y. et al. The molecular basis of tRNA selectivity by human pseudouridine synthase 3. Mol. Cell 84, 2472–2489.e2478 (2024).
Deogharia, M., Mukhopadhyay, S., Joardar, A. & Gupta, R. The human ortholog of archaeal Pus10 produces pseudouridine 54 in select tRNAs where its recognition sequence contains a modified residue. RNA 25, 336–351 (2019).
Mukhopadhyay, S., Deogharia, M. & Gupta, R. Mammalian nuclear TRUB1, mitochondrial TRUB2, and cytoplasmic PUS10 produce conserved pseudouridine 55 in different sets of tRNA. RNA 27, 66–79 (2021).
Antonicka, H. et al. A pseudouridine synthase module is essential for mitochondrial protein synthesis and cell viability. Embo Rep. 18, 28–38 (2017).
Xu, H. et al. A comprehensive tRNA pseudouridine map uncovers targets dependent on human stand-alone pseudouridine synthases. Nat. Cell Biol. 27, 2186–2197 (2025).
Wu, G. et al. The TOR signaling pathway regulates starvation-induced pseudouridylation of yeast U2 snRNA. RNA 22, 1146–1152 (2016).
Li, J., Zhu, W. Y., Yang, W. Q., Li, C. T. & Liu, R. J. The occurrence order and cross-talk of different tRNA modifications. Sci. China Life Sci. 64, 1423–1436 (2021).
Ishida, K. et al. Pseudouridine at position 55 in tRNA controls the contents of other modified nucleotides for low-temperature adaptation in the extreme-thermophilic eubacterium Thermus thermophilus. Nucleic Acids Res. 39, 2304–2318 (2011).
Yared, M. J., Marcelot, A. & Barraud, P. Beyond the anticodon: tRNA core modifications and their impact on structure, translation and stress adaptation. Genes 15, 374 (2024).
Porat, J. Circuit logic: interdependent RNA modifications shape mRNA and noncoding RNA structure and function. RNA 31, 613–622 (2025).
Guegueniat, J. et al. The human pseudouridine synthase PUS7 recognizes RNA with an extended multi-domain binding surface. Nucleic Acids Res. 49, 11810–11822 (2021).
Czudnochowski, N., Wang, A. L., Finer-Moore, J. & Stroud, R. M. In human pseudouridine synthase 1 (hPus1), a C-terminal helical insert blocks tRNA from binding in the same orientation as in the Pus1 bacterial homologue TruA, consistent with their different target selectivities. J. Mol. Biol. 425, 3875–3887 (2013).
Hoang, C. & Ferre-D’Amare, A. R. Cocrystal structure of a tRNA Psi55 pseudouridine synthase: nucleotide flipping by an RNA-modifying enzyme. Cell 107, 929–939 (2001).
Yarian, C. S. et al. Structural and functional roles of the N1- and N3-protons of psi at tRNA’s position 39. Nucleic Acids Res. 27, 3543–3549 (1999).
Rintala-Dempsey, A. C. & Kothe, U. Eukaryotic stand-alone pseudouridine synthases - RNA modifying enzymes and emerging regulators of gene expression?. RNA Biol. 14, 1185–1196 (2017).
Agris, P. F. Bringing order to translation: the contributions of transfer RNA anticodon-domain modifications. Embo Rep 9, 629–635 (2008).
Lei, Z. & Yi, C. A Radiolabeling-free, qPCR-based method for locus-specific pseudouridine detection. Angew. Chem. Int. Ed. Engl. 56, 14878–14882 (2017).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Van Nostrand, E. L. et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat. Methods 13, 508–514 (2016).
Martin, M. Cutadapt removes adapter sequences from high-through sequencing reads. EMBnet J. 17, 10–12 (2011).
Shen, W., Sipos, B. & Zhao, L. SeqKit2: a Swiss army knife for sequence and alignment processing. Imeta 3, e191 (2024).
Smith, T., Heger, A. & Sudbery, I. UMI-tools: modeling sequencing errors in unique molecular identifiers to improve quantification accuracy. Genome Res. 27, 491–499 (2017).
Chan, P. P. & Lowe, T. M. GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 44, D184–D189 (2016).
Danecek, P. et al. Twelve years of SAMtools and BCFtools. Gigascience 10, giab008 (2021).
Kim, D., Paggi, J. M., Park, C., Bennett, C. & Salzberg, S. L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915 (2019).
Gruber, A. R., Lorenz, R., Bernhart, S. H., Neubock, R. & Hofacker, I. L. The Vienna RNA websuite. Nucleic Acids Res. 36, W70–W74 (2008).
Wenqing, L. PsiUInterplayAnalysis. Zenodo https://doi.org/10.5281/zenodo.18205172 (2026).
Acknowledgements
The authors would like to thank Dr. Xiaoting Zhang, Dr. Hanxiao Sun, Dr. Jinmin Yang, and Jinghan Zhou for discussions, and Dr. Hongxia Chen for providing the PUS7L knockout cell line. This work was supported by the National Key R&D Program of China (2021YFC2302400 to M.Z. and 2023YFC3402200 to C.Y.), the National Natural Science Foundation of China (22425071 to C.Y. and 61575008 to M.Z.), the Beijing National Science Foundation of China (Z231100002723005 to C.Y.), and the Natural Science Foundation of Sichuan Province of China (2026NSFSC0869 to B.H.). This work was supported by the New Cornerstone Science Foundation through the XPLORER PRIZE. This work was supported by the Open Research Fund of the National Center for Protein Sciences at Peking University and the Chinese Universities Scientific Fund 2025TC012.
Author information
Authors and Affiliations
Contributions
C.Y. and M.Z. conceived the project and designed the experiments; M.Z., C.Y., and W.L. wrote the manuscript; M.Z. performed the experiments with the help of L.W. and Y.D.; W.L. and Y.M. designed and performed the bioinformatics analysis for NGS samples with the help of B.H. B.L. constructed the PUS1L and three RPUSD knockout cell lines. Y.Z. performed the immunofluorescence assays.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Tsutomu (Tom) Suzuki, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Liu, W., Ma, Y., Wang, L. et al. Quantitative analysis of small RNA pseudouridylation reveals interplay of PUS enzymes in tRNA anticodon stem-loop. Nat Commun (2026). https://doi.org/10.1038/s41467-026-69177-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-026-69177-7