Abstract
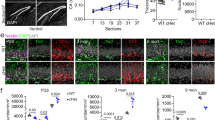
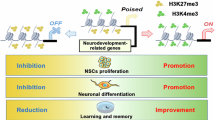
Methylation of lysine 4 on histone H3 (H3K4) is enriched on active promoters and enhancers where it promotes gene activation. Disruption of H3K4 methylation is associated with numerous neurodevelopmental diseases (NDDs) that display intellectual disability and abnormal body growth. Here, we perturb H3K4 methylation in the medial ganglionic eminence (MGE) and hypothalamus, two brain regions associated with these disease phenotypes. These mutant mice have fewer forebrain interneurons, deficient network rhythmogenesis, and increased spontaneous seizures and seizure susceptibility. Mutant mice are significantly smaller than control littermates, but they eventually became obese due to striking changes in the genetic and cellular hypothalamus environment in these mice. Perturbation of H3K4 methylation in these cells produces deficits in numerous NDD-associated behaviors, with a bias for more severe phenotypes in female mice. Single nuclei sequencing reveals transcriptional changes in the embryonic and adult brain that underlie many of these phenotypes. In sum, our findings highlight the critical role of H3K4 methylation in regulating survival and cell-specific gene regulatory mechanisms in forebrain GABAergic and hypothalamic cells during neurodevelopment to control network excitability and body size homoeostasis.
Similar content being viewed by others
Data availability
The single-cell sequencing data generated in this study have been deposited in the Gene Expression Omnibus (GEO) repository under Superseries accession code GSE293881, which includes our single-cell ATAC-seq (GSE293655) and single-cell RNA-seq (GSE293751) datasets, all of which are publicly available. Details on cell counts, number of animals used per experiment, and analyses of differentially expressed genes for all comparisons described in the manuscript are provided in the Supplementary Data files. Additional data & information used in this study are available at Synapse.org. For any additional inquiries about data accessibility and analysis, please email tim.petros@nih.gov.
References
Boukas, L. et al. Coexpression patterns define epigenetic regulators associated with neurological dysfunction. Genome Res. 29, 532–542 (2019).
Bjornsson, H. T. The Mendelian disorders of the epigenetic machinery. Genome Res. 25, 1473–1481 (2015).
Tatton-Brown, K. et al. Mutations in epigenetic regulation genes are a major cause of overgrowth with intellectual disability. Am. J. Hum. Genet. 100, 725–736 (2017).
Fahrner, J. A. & Bjornsson, H. T. Mendelian disorders of the epigenetic machinery: postnatal malleability and therapeutic prospects. Hum. Mol. Genet. 28, R254–R264 (2019).
Strahl, B. D. & Allis, C. D. The language of covalent histone modifications. Nature 403, 41–45 (2000).
Ritchie, F. D. & Lizarraga, S. B. The role of histone methyltransferases in neurocognitive disorders associated with brain size abnormalities. Front. Neurosci. 17, 989109 (2023).
Shen, E., Shulha, H., Weng, Z. & Akbarian, S. Regulation of histone H3K4 methylation in brain development and disease. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369, 20130514 (2014).
Collins, B. E., Greer, C. B., Coleman, B. C. & Sweatt, J. D. Histone H3 lysine K4 methylation and its role in learning and memory. Epigenetics Chromatin 12, 7 (2019).
Vallianatos, C. N. & Iwase, S. Disrupted intricacy of histone H3K4 methylation in neurodevelopmental disorders. Epigenomics 7, 503–519 (2015).
Maze, I. et al. Critical Role of Histone Turnover in Neuronal Transcription and Plasticity. Neuron 87, 77–94 (2015).
Xia, W. & Jiao, J. Histone variant H3.3 orchestrates neural stem cell differentiation in the developing brain. Cell Death Differ. 24, 1548–1563 (2017).
Funk, O. H., Qalieh, Y., Doyle, D. Z., Lam, M. M. & Kwan, K. Y. Postmitotic accumulation of histone variant H3.3 in new cortical neurons establishes neuronal chromatin, transcriptome, and identity. Proc. Natl. Acad. Sci. USA 119, e2116956119 (2022).
Klein, R. H. & Knoepfler, P. S. Knockout tales: the versatile roles of histone H3.3 in development and disease. Epigenetics Chromatin 16, 38 (2023).
Van, H. T., Xie, G., Dong, P., Liu, Z. & Ge, K. KMT2 Family of H3K4 methyltransferases: enzymatic activity-dependent and -independent functions. J. Mol. Biol. 436, 168453 (2024).
Ng, S. B. et al. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat. Genet. 42, 790–793 (2010).
Jones, W. D. et al. De novo mutations in MLL cause Wiedemann-Steiner syndrome. Am. J. Hum. Genet. 91, 358–364 (2012).
Strom, S. P. et al. De Novo variants in the KMT2A (MLL) gene causing atypical Wiedemann-Steiner syndrome in two unrelated individuals identified by clinical exome sequencing. BMC Med. Genet. 15, 49 (2014).
Siano, M. A. et al. De novo mutation in KMT2C manifesting as Kleefstra syndrome 2: case report and literature review. Pediatr. Rep. 14, 131–139 (2022).
Lim, D. A. et al. Chromatin remodelling factor Mll1 is essential for neurogenesis from postnatal neural stem cells. Nature 458, 529–533 (2009).
Kerimoglu, C. et al. KMT2A and KMT2B Mediate memory function by affecting distinct genomic regions. Cell Rep. 20, 538–548 (2017).
Michurina, A. et al. Postnatal expression of the lysine methyltransferase SETD1B is essential for learning and the regulation of neuron-enriched genes. EMBO J. 41, e106459 (2022).
Nagahama, K. et al. Setd1a insufficiency in mice attenuates excitatory synaptic function and recapitulates schizophrenia-related behavioral abnormalities. Cell Rep. 32, 108126 (2020).
Qin, L. et al. Deficiency of autism risk factor ASH1L in prefrontal cortex induces epigenetic aberrations and seizures. Nat. Commun. 12, 6589 (2021).
Bandler, R. C., Mayer, C. & Fishell, G. Cortical interneuron specification: the juncture of genes, time and geometry. Curr. Opin. Neurobiol. 42, 17–24 (2017).
Williams, R. H. & Riedemann, T. Development, diversity, and death of MGE-derived cortical interneurons. Int. J. Mol. Sci. 22, https://doi.org/10.3390/ijms22179297 (2021).
Ferrer, C. & De Marco Garcia, N. V. The role of inhibitory interneurons in circuit assembly and refinement across sensory cortices. Front. Neural Circuits 16, 866999 (2022).
Bozzi, Y., Casarosa, S. & Caleo, M. Epilepsy as a neurodevelopmental disorder. Front. Psychiatry 3, 19 (2012).
Inan, M., Petros, T. J. & Anderson, S. A. Losing your inhibition: linking cortical GABAergic interneurons to schizophrenia. Neurobiol. Dis. 53, 36–48 (2013).
Takano, T. Interneuron dysfunction in syndromic autism: recent advances. Dev. Neurosci. 37, 467–475 (2015).
Katsarou, A. M., Moshe, S. L. & Galanopoulou, A. S. Interneuronopathies and their role in early life epilepsies and neurodevelopmental disorders. Epilepsia Open 2, 284–306 (2017).
Batista-Brito, R., Machold, R., Klein, C. & Fishell, G. Gene expression in cortical interneuron precursors is prescient of their mature function. Cereb Cortex 18, 2306–2317 (2008).
Parikshak, N. N. et al. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell 155, 1008–1021 (2013).
Schork, A. J. et al. A genome-wide association study of shared risk across psychiatric disorders implicates gene regulation during fetal neurodevelopment. Nat. Neurosci. 22, 353–361 (2019).
Trevino, A. E. et al. Chromatin accessibility dynamics in a model of human forebrain development. Science 367, https://doi.org/10.1126/science.aay1645 (2020).
Paulsen, B. et al. Autism genes converge on asynchronous development of shared neuron classes. Nature 602, 268–273 (2022).
Villa, C. E. et al. CHD8 haploinsufficiency links autism to transient alterations in excitatory and inhibitory trajectories. Cell Rep. 39, 110615 (2022).
Liu, D. et al. Impact of schizophrenia GWAS loci converge onto distinct pathways in cortical interneurons vs glutamatergic neurons during development. Mol. Psychiatry 27, 4218–4233 (2022).
Jang, Y. et al. H3.3K4M destabilizes enhancer H3K4 methyltransferases MLL3/MLL4 and impairs adipose tissue development. Nucleic Acids Res. 47, 607–620 (2019).
Sussel, L., Marin, O., Kimura, S. & Rubenstein, J. L. Loss of Nkx2.1 homeobox gene function results in a ventral to dorsal molecular respecification within the basal telencephalon: evidence for a transformation of the pallidum into the striatum. Development 126, 3359–3370 (1999).
Sandberg, M. et al. Transcriptional Networks Controlled by NKX2-1 in the Development of Forebrain GABAergic Neurons. Neuron 91, 1260–1275 (2016).
Xu, Q., Tam, M. & Anderson, S. A. Fate mapping Nkx2.1-lineage cells in the mouse telencephalon. J. Comp. Neurol. 506, 16–29 (2008).
Orquera, D. P. et al. The homeodomain transcription factor NKX2.1 is essential for the early specification of melanocortin neuron identity and activates Pomc expression in the developing hypothalamus. J. Neurosci. 39, 4023–4035 (2019).
Ma, T., Wong, S. Z. H., Lee, B., Ming, G. L. & Song, H. Decoding neuronal composition and ontogeny of individual hypothalamic nuclei. Neuron 109, 1150–1167 (2021).
Herrera Moro Chao, D. et al. Hypothalamic astrocytes control systemic glucose metabolism and energy balance. Cell Metab. 34, 1532–1547 (2022).
Zhang, Y. H. et al. Cascade diversification directs generation of neuronal diversity in the hypothalamus. Cell Stem Cell 28, 1483–1499 (2021).
Commerford, S. L., Carsten, A. L. & Cronkite, E. P. Histone turnover within nonproliferating cells. Proc. Natl. Acad. Sci. USA 79, 1163–1165 (1982).
Wenderski, W. & Maze, I. Histone turnover and chromatin accessibility: Critical mediators of neurological development, plasticity, and disease. Bioessays 38, 410–419 (2016).
Rhodes, C. T. et al. Loss of Ezh2 in the medial ganglionic eminence alters interneuron fate, cell morphology and gene expression profiles. Front. Cell Neurosci. 18, 1334244 (2024).
Liodis, P. et al. Lhx6 activity is required for the normal migration and specification of cortical interneuron subtypes. J. Neurosci. 27, 3078–3089 (2007).
Van Erum, J., Van Dam, D. & De Deyn, P. P. PTZ-induced seizures in mice require a revised Racine scale. Epilepsy Behav. 95, 51–55 (2019).
Medina, A. E., Manhaes, A. C. & Schmidt, S. L. Sex differences in sensitivity to seizures elicited by pentylenetetrazol in mice. Pharmacol. Biochem. Behav. 68, 591–596 (2001).
Fisahn, A., Pike, F. G., Buhl, E. H. & Paulsen, O. Cholinergic induction of network oscillations at 40 Hz in the hippocampus in vitro. Nature 394, 186–189 (1998).
Mann, E. O., Suckling, J. M., Hajos, N., Greenfield, S. A. & Paulsen, O. Perisomatic feedback inhibition underlies cholinergically induced fast network oscillations in the rat hippocampus in vitro. Neuron 45, 105–117 (2005).
Colgin, L. L. & Moser, E. I. Gamma oscillations in the hippocampus. Physiology 25, 319–329 (2010).
Duzel, E., Penny, W. D. & Burgess, N. Brain oscillations and memory. Curr. Opin. Neurobiol. 20, 143–149 (2010).
Buzsaki, G. & Silva, F. L. High frequency oscillations in the intact brain. Prog. Neurobiol. 98, 241–249 (2012).
Drexel, M. et al. Selective silencing of hippocampal parvalbumin interneurons induces development of recurrent spontaneous limbic seizures in mice. J. Neurosci. 37, 8166–8179 (2017).
Andrioli, A., Alonso-Nanclares, L., Arellano, J. I. & DeFelipe, J. Quantitative analysis of parvalbumin-immunoreactive cells in the human epileptic hippocampus. Neuroscience 149, 131–143 (2007).
Andre, V., Marescaux, C., Nehlig, A. & Fritschy, J. M. Alterations of hippocampal GAbaergic system contribute to development of spontaneous recurrent seizures in the rat lithium-pilocarpine model of temporal lobe epilepsy. Hippocampus 11, 452–468 (2001).
Honey, C. J. & Valiante, T. Neuroscience: when a single image can cause a seizure. Curr. Biol. 27, R394–R397 (2017).
Southwell, D. G. et al. Intrinsically determined cell death of developing cortical interneurons. Nature 491, 109–113 (2012).
Denaxa, M. et al. Modulation of apoptosis controls inhibitory interneuron number in the cortex. Cell Rep. 22, 1710–1721 (2018).
Priya, R. et al. Activity regulates cell death within cortical interneurons through a calcineurin-dependent mechanism. Cell Rep. 22, 1695–1709 (2018).
Winstanley, C. A., Eagle, D. M. & Robbins, T. W. Behavioral models of impulsivity in relation to ADHD: translation between clinical and preclinical studies. Clin. Psychol. Rev. 26, 379–395 (2006).
Gao, Y., Aljazi, M. B. & He, J. Kdm6b Haploinsufficiency causes ASD/ADHD-like behavioral deficits in mice. Front. Behav. Neurosci. 16, 905783 (2022).
Ioannidou, C., Marsicano, G. & Busquets-Garcia, A. Assessing prepulse inhibition of startle in mice. Bio. Protoc. 8, e2789 (2018).
Heller, H. C. et al. Nest building is impaired in the Ts65Dn mouse model of Down syndrome and rescued by blocking 5HT2a receptors. Neurobiol. Learn Mem. 116, 162–171 (2014).
Jirkof, P. Burrowing and nest building behavior as indicators of well-being in mice. J. Neurosci. Methods 234, 139–146 (2014).
Mayer, C. et al. Developmental diversification of cortical inhibitory interneurons. Nature 555, 457–462 (2018).
Pai, E. L. et al. Maf and Mafb control mouse pallial interneuron fate and maturation through neuropsychiatric disease gene regulation. Elife 9, https://doi.org/10.7554/elife.54903 (2020).
Wu, S. J. et al. Pyramidal neurons proportionately alter the identity and survival of specific cortical interneuron subtypes. Nature https://doi.org/10.1038/s41586-025-09996-8 (2026).
Langfelder, P. & Horvath, S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinform. 9, 559 (2008).
Morabito, S., Reese, F., Rahimzadeh, N., Miyoshi, E. & Swarup, V. hdWGCNA identifies co-expression networks in high-dimensional transcriptomics data. Cell Rep. Methods 3, 100498 (2023).
Du, J., Zhang, L., Weiser, M., Rudy, B. & McBain, C. J. Developmental expression and functional characterization of the potassium-channel subunit Kv3.1b in parvalbumin-containing interneurons of the rat hippocampus. J. Neurosci. 16, 506–518 (1996).
Lau, D. et al. Impaired fast-spiking, suppressed cortical inhibition, and increased susceptibility to seizures in mice lacking Kv3.2 K+ channel proteins. J. Neurosci. 20, 9071–9085 (2000).
Wu, S. J. et al. Cortical somatostatin interneuron subtypes form cell-type-specific circuits. Neuron 111, 2675–2692.e9 (2023).
Shimogori, T. et al. A genomic atlas of mouse hypothalamic development. Nat. Neurosci. 13, 767–775 (2010).
Kim, D. W. et al. The cellular and molecular landscape of hypothalamic patterning and differentiation from embryonic to late postnatal development. Nat. Commun. 11, 4360 (2020).
Zhou, X. et al. Cellular and molecular properties of neural progenitors in the developing mammalian hypothalamus. Nat. Commun. 11, 4063 (2020).
Kim, D. W. et al. Decoding gene networks controlling hypothalamic and prethalamic neuron development. Cell Rep. 44, 115858 (2025).
van Velthoven, C. T. J. et al. Transcriptomic and spatial organization of telencephalic GABAergic neurons. Nature 647, 143–156 (2025).
Jais, A. & Bruning, J. C. Arcuate nucleus-dependent rRegulation of metabolism-pathways to obesity and diabetes mellitus. Endocr. Rev. 43, 314–328 (2022).
Khodai, T. & Luckman, S. M. Ventromedial nucleus of the hypothalamus neurons under the magnifying glass. Endocrinology 162, https://doi.org/10.1210/endocr/bqab141 (2021).
Steuernagel, L. et al. HypoMap-a unified single-cell gene expression atlas of the murine hypothalamus. Nat. Metab. 4, 1402–1419 (2022).
Horvath, T. L. et al. Synaptic input organization of the melanocortin system predicts diet-induced hypothalamic reactive gliosis and obesity. Proc. Natl. Acad. Sci. USA 107, 14875–14880 (2010).
Chowen, J. A. et al. The role of astrocytes in the hypothalamic response and adaptation to metabolic signals. Prog. Neurobiol. 144, 68–87 (2016).
Gonzalez-Garcia, I. & Garcia-Caceres, C. Hypothalamic astrocytes as a specialized and responsive cell population in obesity. Int. J. Mol. Sci. 22, (2021).
Verkhratsky, A. & Nedergaard, M. Physiology of astroglia. Physiol. Rev. 98, 239–389 (2018).
Lyon, K. A. & Allen, N. J. From Synapses to Circuits, Astrocytes Regulate Behavior. Front. Neural Circuits 15, 786293 (2021).
Goodman, T. & Hajihosseini, M. K. Hypothalamic tanycytes-masters and servants of metabolic, neuroendocrine, and neurogenic functions. Front. Neurosci. 9, 387 (2015).
Campbell, J. N. et al. A molecular census of arcuate hypothalamus and median eminence cell types. Nat. Neurosci. 20, 484–496 (2017).
Robins, S. C. et al. alpha-Tanycytes of the adult hypothalamic third ventricle include distinct populations of FGF-responsive neural progenitors. Nat. Commun. 4, 2049 (2013).
Buller, S. et al. Median eminence myelin continuously turns over in adult mice. Mol. Metab. 69, 101690 (2023).
Stengel, A. & Tache, Y. Central somatostatin signaling and regulation of food intake. Ann. N. Y. Acad. Sci. 1455, 98–104 (2019).
Chen, R., Wu, X., Jiang, L. & Zhang, Y. Single-cell RNA-seq reveals hypothalamic cell diversity. Cell Rep. 18, 3227–3241 (2017).
Esteve, N. A. et al. Tanycyte radial morphology and proliferation are influenced by fibroblast growth factor receptor 1 and high-fat diet. Eur. J. Neurosci. 60, 5000–5018 (2024).
Geller, S. et al. Tanycytes regulate Lipid Homeostasis by sensing free fatty acids and signaling to key hypothalamic neuronal populations via FGF21 secretion. Cell Metab. 30, 833–844.e7 (2019).
Kaminskas, B. et al. Characterisation of endogenous players in fibroblast growth factor-regulated functions of hypothalamic tanycytes and energy-balance nuclei. J. Neuroendocrinol. 31, e12750 (2019).
Bentsen, M. A. et al. Transcriptomic analysis links diverse hypothalamic cell types to fibroblast growth factor 1-induced sustained diabetes remission. Nat. Commun. 11, 4458 (2020).
Serdyukova, K. et al. Leveraging dominant-negative histone H3 K-to-M mutations to study chromatin during differentiation and development. Development 150, https://doi.org/10.1242/dev.202169 (2023).
Vilchez-Acosta, A. et al. Specific contribution of Reelin expressed by Cajal-Retzius cells or GABAergic interneurons to cortical lamination. Proc. Natl. Acad. Sci. USA 119, e2120079119 (2022).
Reichard, J. et al. DNMT1-mediated regulation of somatostatin-positive interneuron migration impacts cortical architecture and function. Nat. Commun. 16, 6834 (2025).
Doischer, D. et al. Postnatal differentiation of basket cells from slow to fast signaling devices. J. Neurosci. 28, 12956–12968 (2008).
Caccavano, A. P. et al. Divergent opioid-mediated suppression of inhibition between hippocampus and neocortex across species and development. Neuron 113, 1805–1822.e7 (2025).
Cardin, J. A. et al. Driving fast-spiking cells induces gamma rhythm and controls sensory responses. Nature 459, 663–667 (2009).
Cammarota, M., Losi, G., Chiavegato, A., Zonta, M. & Carmignoto, G. Fast spiking interneuron control of seizure propagation in a cortical slice model of focal epilepsy. J. Physiol. 591, 807–822 (2013).
Zheng, Y. & Chen, J. Voltage-gated potassium channels and genetic epilepsy. Front. Neurol. 15, 1466075 (2024).
van Luijtelaar, G. & Sitnikova, E. Global and focal aspects of absence epilepsy: the contribution of genetic models. Neurosci. Biobehav. Rev. 30, 983–1003 (2006).
Harvey, M., Lau, D., Civillico, E., Rudy, B. & Contreras, D. Impaired long-range synchronization of gamma oscillations in the neocortex of a mouse lacking Kv3.2 potassium channels. J. Neurophysiol. 108, 827–833 (2012).
Andrade-Talavera, Y., Arroyo-Garcia, L. E., Chen, G., Johansson, J. & Fisahn, A. Modulation of Kv3.1/Kv3.2 promotes gamma oscillations by rescuing Abeta-induced desynchronization of fast-spiking interneuron firing in an AD mouse model in vitro. J. Physiol. 598, 3711–3725 (2020).
Khazaei, S. et al. Single substitution in H3.3G34 alters DNMT3A recruitment to cause progressive neurodegeneration. Cell 186, 1162–1178.e20 (2023).
Mohammad, F. & Helin, K. Oncohistones: drivers of pediatric cancers. Genes Dev. 31, 2313–2324 (2017).
Bannister, A. J. & Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 21, 381–395 (2011).
Kundakovic, M. & Tickerhoof, M. Epigenetic mechanisms underlying sex differences in the brain and behavior. Trends Neurosci. 47, 18–35 (2024).
Forneris, F., Binda, C., Adamo, A., Battaglioli, E. & Mattevi, A. Structural basis of LSD1-CoREST selectivity in histone H3 recognition. J. Biol. Chem. 282, 20070–20074 (2007).
Yagi, M. et al. Bivalent chromatin instructs lineage specification during hematopoiesis. Cell 188, 4314–4331.e29 (2025).
Adam, M. P. et al. Kabuki syndrome: international consensus diagnostic criteria. J. Med. Genet. 56, 89–95 (2019).
Jung, Y. L., Hung, C., Choi, J., Lee, E. A. & Bodamer, O. Characterizing the molecular impact of KMT2D variants on the epigenetic and transcriptional landscapes in Kabuki syndrome. Hum. Mol. Genet. 32, 2251–2261 (2023).
Huisman, C. et al. Single cell transcriptome analysis of developing arcuate nucleus neurons uncovers their key developmental regulators. Nat. Commun. 10, 3696 (2019).
Timper, K. & Bruning, J. C. Hypothalamic circuits regulating appetite and energy homeostasis: pathways to obesity. Dis. Model Mech. 10, 679–689 (2017).
Pasquettaz, R. et al. Peculiar protrusions along tanycyte processes face diverse neural and nonneural cell types in the hypothalamic parenchyma. J. Comp .Neurol. 529, 553–575 (2021).
Dali, R., Estrada-Meza, J. & Langlet, F. Tanycyte, the neuron whisperer. Physiol. Behav. 263, 114108 (2023).
Qin, C., Li, J. & Tang, K. The paraventricular nucleus of the hypothalamus: development, function, and human diseases. Endocrinology 159, 3458–3472 (2018).
Mendoza-Herrera, K. et al. The Leptin system and diet: a mini review of the current evidence. Front. Endocrinol. 12, 749050 (2021).
Frederich, R. C. et al. Leptin levels reflect body lipid content in mice: evidence for diet-induced resistance to leptin action. Nat. Med. 1, 1311–1314 (1995).
Garcia-Caceres, C. et al. Role of astrocytes, microglia, and tanycytes in brain control of systemic metabolism. Nat. Neurosci. 22, 7–14 (2019).
Mukai, J. et al. Recapitulation and reversal of schizophrenia-related phenotypes in setd1a-deficient mice. Neuron 104, 471–487 (2019).
Bolte, S. et al. Sex and gender in neurodevelopmental conditions. Nat. Rev. Neurol. 19, 136–159 (2023).
Seeman, M. V. Psychopathology in women and men: focus on female hormones. Am. J. Psychiatry 154, 1641–1647 (1997).
Zahn-Waxler, C., Shirtcliff, E. A. & Marceau, K. Disorders of childhood and adolescence: gender and psychopathology. Annu. Rev. Clin. Psychol. 4, 275–303 (2008).
Shen, E. Y. et al. Epigenetics and sex differences in the brain: A genome-wide comparison of histone-3 lysine-4 trimethylation (H3K4me3) in male and female mice. Exp. Neurol. 268, 21–29 (2015).
Hophing, L., Kyriakopoulos, P. & Bui, E. Sex and gender differences in epilepsy. Int. Rev. Neurobiol. 164, 235–276 (2022).
Akman, O., Moshe, S. L. & Galanopoulou, A. S. Sex-specific consequences of early life seizures. Neurobiol. Dis. 72, 153–166 (2014).
Clemens, A. M. et al. Estrus-Cycle Regulation of Cortical Inhibition. Curr. Biol. 29, 605–615.e6 (2019).
Aleman, A., Kahn, R. S. & Selten, J. P. Sex differences in the risk of schizophrenia: evidence from meta-analysis. Arch. Gen. Psychiatry 60, 565–571 (2003).
Santangelo, S. L. et al. Tourette’s syndrome: what are the influences of gender and comorbid obsessive-compulsive disorder? J. Am. Acad. Child Adolesc. Psychiatry 33, 795–804 (1994).
Solberg, B. S. et al. Gender differences in psychiatric comorbidity: a population-based study of 40 000 adults with attention deficit hyperactivity disorder. Acta Psychiatr. Scand. 137, 176–186 (2018).
Correa, S. M. et al. An estrogen-responsive module in the ventromedial hypothalamus selectively drives sex-specific activity in females. Cell Rep. 10, 62–74 (2015).
Giedd, J. N., Castellanos, F. X., Rajapakse, J. C., Vaituzis, A. C. & Rapoport, J. L. Sexual dimorphism of the developing human brain. Prog Neuropsychopharmacol. Biol. Psychiatry 21, 1185–1201 (1997).
Rijpkema, M. et al. Normal sexual dimorphism in the human basal ganglia. Hum. Brain Mapp. 33, 1246–1252 (2012).
Van Zandt, M. & Pittenger, C. Sex differences in histamine regulation of striatal dopamine. J. Neurosci. 45, e2182242025 (2025).
Shah, N. M. et al. Visualizing sexual dimorphism in the brain. Neuron 43, 313–319 (2004).
Brinkman, A. B. et al. Histone modification patterns associated with the human X chromosome. EMBO Rep. 7, 628–634 (2006).
Mo, R., Rao, S. M. & Zhu, Y. J. Identification of the MLL2 complex as a coactivator for estrogen receptor alpha. J. Biol. Chem. 281, 15714–15720 (2006).
Yevoo, P. E., Fontanini, A. & Maffei, A. Modulation of sweet preference by neurosteroid-sensitive, delta-GABA(A) receptors in adult mouse gustatory insular cortex. Curr. Biol. 35, 1047–1060 (2025).
Fogarty, M. et al. Spatial genetic patterning of the embryonic neuroepithelium generates GABAergic interneuron diversity in the adult cortex. J. Neurosci. 27, 10935–10946 (2007).
Madisen, L. et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140 (2010).
Mo, A. et al. Epigenomic signatures of neuronal diversity in the mammalian brain. Neuron 86, 1369–1384 (2015).
Arroyo-Garcia, L. E. et al. Impaired spike-gamma coupling of area CA3 fast-spiking interneurons as the earliest functional impairment in the App(NL-G-F) mouse model of Alzheimer’s disease. Mol. Psychiatry 26, 5557–5567 (2021).
Pizzirusso, G., Sundstrom, S. & Arroyo-Garcia, L. E. Efficient, automatic, and reproducible patch clamp data analysis with “Auto ANT”, a user-friendly interface for batch analysis of patch clamp recordings. Neuroinformatics 23, 24 (2025).
Shimada, T. & Yamagata, K. Pentylenetetrazole-Induced Kindling Mouse Model. J. Vis. Exp. 136, https://doi.org/10.3791/56573 (2018).
Lee, D. R., Zhang, Y., Rhodes, C. T. & Petros, T. J. Generation of single-cell and single-nuclei suspensions from embryonic and adult mouse brains. STAR Protoc. 4, 101944 (2023).
Satija, R., Farrell, J. A., Gennert, D., Schier, A. F. & Regev, A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502 (2015).
Germain, P. L., Lun, A., Garcia Meixide, C., Macnair, W. & Robinson, M. D. Doublet identification in single-cell sequencing data using scDblFinder. F1000Res. 10, 979 (2021).
Stuart, T., Srivastava, A., Madad, S., Lareau, C. A. & Satija, R. Single-cell chromatin state analysis with Signac. Nat. Methods 18, 1333–1341 (2021).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587 (2021).
Yao, Z. et al. A taxonomy of transcriptomic cell types across the isocortex and hippocampal formation. Cell 184, 3222–3241 (2021).
Xu, S. et al. Using clusterProfiler to characterize multiomics data. Nat. Protoc. 19, 3292–3320 (2024).
Cao, J. et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502 (2019).
Acknowledgements
We thank all members of the Section on Cellular and Molecular Neurodevelopment, as well as Pedro Rocha, Ariel Levine and Kai Ge for discussion and comments on this project and manuscript. We thank Kai Ge (NIDDK) for the LSL-K4M mice. We thank the NICHD Molecular Genomics Core, specifically Fabio Faucz, Vivek Mahadevan, Tianwei Li and James Iben; the NIDDK Mouse Metabolism Core, particularly Oksana Gavrilova and Naili Liu, for assistance with measuring body composition; the NINDS and NICHD animal facility for mouse husbandry assistance. This work utilized the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov). This work was supported, in part, by the NIMH IRP Rodent Behavioral Core (MH002952). This project was funded by NICHD intramural projects HD008962 (T.J.P.), HD008986 (R.K.D.), HD001205 (C.J.M.); NICHD Scientific Director’s Award (T.J.P); NICHD Intramural Research Fellowship (J.L.); NICHD Career Development Award (J.L.). This research was supported by the Intramural Research Program of the National Institutes of Health (NIH). The contributions of the NIH author(s) are considered Works of the United States Government. The findings and conclusions presented in this paper are those of the author(s) and do not necessarily reflect the views of the NIH or the U.S. Department of Health and Human Services.
Funding
Open access funding provided by the National Institutes of Health.
Author information
Authors and Affiliations
Contributions
Conceptualization – J.L. and T.J.P.; Investigation – J.L., A.F.T., G.P., A.C., R.C., D.A., Y.Z., and K.A.P.; Formal analysis – J.L., G.P., K.A.P., and M.S.; Software – M.S.; Supervision – R.K.D., C.J.M., and T.J.P.; Funding acquisition – J.L., R.K.D., C.J.M., and T.J.P.; Writing, original draft – J.L. and T.J.P.; Writing, review & editing – all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Daniel Vogt and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, J., Tanzillo, A.F., Pizzirusso, G. et al. Reducing methylation of histone 3.3 lysine 4 in the medial ganglionic eminence and hypothalamus recapitulates neurodevelopmental disorder phenotypes. Nat Commun (2026). https://doi.org/10.1038/s41467-026-69248-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-026-69248-9