Abstract
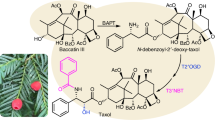
Paclitaxel rapidly became one of the most effective anticancer drugs. However, the production of paclitaxel is hindered by substantial challenges, particularly considering the significant quantities of drug required and the inherently low concentration of paclitaxel and its intermediates in plants. Paclitaxel is currently produced in a so-called semi-synthesis in which baccatin III is extracted from Taxus species and chemically converted to paclitaxel. Despite the fact that many of the intermediates of paclitaxel biosynthesis are yet to be experimentally determined, a set of recent papers—facilitated by the sequencing and assembly of three Taxus genomes—has uncovered the minimal gene sets for both baccatin III and paclitaxel biosynthesis. Here we summarize the key milestones towards our understanding of paclitaxel biosynthesis and highlight recent advancements made possible by genome-level analysis of potential key genes involved. We argue that these studies will ultimately pave the way towards the elucidation of the entire paclitaxel biosynthetic pathway and facilitate the industrial production of paclitaxel via synthetic biology approaches. However, several major challenges lie ahead before we can fully tap into the amazing curative potential that taxanes provide.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Wani, M. C., Taylor, H. L., Wall, M. E., Coggon, P. & McPhail, A. T. Plant antitumor agents. VI. The isolation and structure of taxol, a novel antileukemic and antitumor agent from Taxus brevifolia. J. Am. Chem. Soc. 93, 2325–2327 (1971).
Huang, C. H., Kingston, D. G., Magri, N. F., Samaranayake, G. & Boettner, F. E. New taxanes from Taxus brevifolia, 2. J. Nat. Prod. 49, 665–669 (1986).
Yang, C.-P. H. & Horwitz, S. B. Taxol®: the first microtubule stabilizing agent. Int. J. Mol. Sci. 18, 1733 (2017).
Jordan, M. A. & Wilson, L. Microtubules as a target for anticancer drugs. Nat. Rev. Cancer 4, 253–265 (2004).
Menzin, A. W., King, S. A., Aikins, J. K., Mikuta, J. J. & Rubin, S. C. Taxol (paclitaxel) was approved by FDA for the treatment of patients with recurrent ovarian cancer. Gynecol. Oncol. 54, 103 (1994).
Zhang, Y., Scossa, F. & Fernie, A. R. The genomes of Taxus species unveil novel candidates in the biosynthesis of taxoids. Mol. Plant 14, 1773–1775 (2021).
Bray, F. et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 74, 229–263 (2024).
Li, J. et al. Chloroplastic metabolic engineering coupled with isoprenoid pool enhancement for committed taxanes biosynthesis in Nicotiana benthamiana. Nat. Commun. 10, 4850 (2019).
Nicolaou, K. et al. Total synthesis of taxol. Nature 367, 630–634 (1994).
Min, L. et al. Strategies and lessons learned from total synthesis of taxol. Chem. Rev. 123, 4934–4971 (2023).
Imamura, Y., Takaoka, K., Komori, Y., Nagatomo, M. & Inoue, M. Total synthesis of taxol enabled by inter‐and intramolecular radical coupling reactions. Angew. Chem. 135, e202219114 (2023).
Watanabe, T., Oga, K., Matoba, H., Nagatomo, M. & Inoue, M. Total synthesis of taxol enabled by intermolecular radical coupling and Pd-catalyzed cyclization. J. Am. Chem. Soc. 145, 25894–25902 (2023).
Zhang, S. et al. Research advances in clinical applications, anticancer mechanism, total chemical synthesis, semi-synthesis and biosynthesis of paclitaxel. Molecules 28, 7517 (2023).
Mutanda, I., Li, J., Xu, F. & Wang, Y. Recent advances in metabolic engineering, protein engineering, and transcriptome-guided insights toward synthetic production of taxol. Front. Bioeng. Biotechnol. 9, 632269 (2021).
Cheng, J. et al. Chromosome-level genome of Himalayan yew provides insights into the origin and evolution of the paclitaxel biosynthetic pathway. Mol. Plant 14, 1199–1209 (2021).
Xiong, X. et al. The Taxus genome provides insights into paclitaxel biosynthesis. Nat. Plants 7, 1026–1036 (2021).
Song, C. et al. Taxus yunnanensis genome offers insights into gymnosperm phylogeny and taxol production. Commun. Biol. 4, 1203 (2021).
Zhang, Y. et al. Synthetic biology identifies the minimal gene set required for paclitaxel biosynthesis in a plant chassis. Mol. Plant 16, 1951–1961 (2023).
Jiang, B. et al. Characterization and heterologous reconstitution of Taxus biosynthetic enzymes leading to baccatin III. Science 383, 622–629 (2024).
Liu, J. C.-T., De La Peña, R., Tocol, C. & Sattely, E. S. Reconstitution of early paclitaxel biosynthetic network. Nat. Commun. 15, 1419 (2024).
Yang, C. et al. Biosynthesis of the highly oxygenated tetracyclic core skeleton of Taxol. Nat. Commun. 15, 2339 (2024).
Zhao, Y. et al. Oxetane ring formation in taxol biosynthesis is catalyzed by a bifunctional cytochrome P450 enzyme. J. Am. Chem. Soc. 146, 801–810 (2023).
Lange, B. M. & Conner, C. F. Taxanes and taxoids of the genus Taxus—a comprehensive inventory of chemical diversity. Phytochemistry 190, 112829 (2021).
Narayanan, A. K. & Nagegowda, D. A. Biosynthesis of the triterpenoid withanolides in Withania somnifera. Curr. Opin. Plant Biol. 81, 102576 (2024).
Garza-Garcia, J. J. O. & Qu, Y. Chemical, pharmacological properties and biosynthesis of opioid mitragynine in Mitragyna speciosa (kratom). Curr. Opin. Plant Biol. 81, 102600 (2024).
Chavez, B. G., Dias, S. L. & D’Auria, J. C. The evolution of tropane alkaloids: coca does it differently. Curr. Opin. Plant Biol. 81, 102606 (2024).
Sánchez-Pérez, R. & Neilson, E. H. The case for sporadic cyanogenic glycoside evolution in plants. Curr. Opin. Plant Biol. 81, 102608 (2024).
Bergman, M. E. & Dudareva, N. Plant specialized metabolism: diversity of terpene synthases and their products. Curr. Opin. Plant Biol. 81, 102607 (2024).
Wani, M. C. & Horwitz, S. B. Nature as a remarkable chemist: a personal story of the discovery and development of Taxol. Anticancer Drugs 25, 482–487 (2014).
Walsh, V. & Goodman, J. From taxol to Taxol: the changing identities and ownership of an anti-cancer drug. Med. Anthropol. 21, 307–336 (2002).
Guénard, D., Guéritte-Voegelein, F., Dubois, J. & Potier, P. Structure-activity relationships of Taxol and Taxotere analogues. J. Natl Cancer Inst. Monogr. 15, 79–82 (1993).
Holton, R., Biediger, R. J. & Boatman, D. P. in TAXOL®: Science and Applications (ed. Suffness, M.) Ch. 5 (CRC, 1995).
Koepp, A. E. et al. Cyclization of geranylgeranyl diphosphate to taxa-4(5),11(12)-diene is the committed step of taxol biosynthesis in Pacific yew. J. Biol. Chem. 270, 8686–8690 (1995).
Rohmer, M., Knani, M., Simonin, P., Sutter, B. & Sahm, H. Isoprenoid biosynthesis in bacteria: a novel pathway for the early steps leading to isopentenyl diphosphate. Biochem. J. 295, 517–524 (1993).
Rohmer, M., Seemann, M., Horbach, S., Bringer-Meyer, S. & Sahm, H. Glyceraldehyde 3-phosphate and pyruvate as precursors of isoprenic units in an alternative non-mevalonate pathway for terpenoid biosynthesis. J. Am. Chem. Soc. 118, 2564–2566 (1996).
Rodríguez-Concepción, M. & Boronat, A. Elucidation of the methylerythritol phosphate pathway for isoprenoid biosynthesis in bacteria and plastids: a metabolic milestone achieved through genomics. Plant Physiol. 130, 1079–1089 (2002).
Guerra-Bubb, J., Croteau, R. & Williams, R. M. The early stages of taxol biosynthesis: an interim report on the synthesis and identification of early pathway metabolites. Nat. Prod. Rep. 29, 683–696 (2012).
Long, R. M., Lagisetti, C., Coates, R. M. & Croteau, R. B. Specificity of the N-benzoyl transferase responsible for the last step of Taxol biosynthesis. Arch. Biochem. Biophys. 477, 384–389 (2008).
Williams, D. C. et al. Intramolecular proton transfer in the cyclization of geranylgeranyl diphosphate to the taxadiene precursor of taxol catalyzed by recombinant taxadiene synthase. Chem. Biol. 7, 969–977 (2000).
Wheeler, A. L. et al. Taxol biosynthesis: differential transformations of taxadien-5 alpha-ol and its acetate ester by cytochrome P450 hydroxylases from Taxus suspension cells. Arch. Biochem. Biophys. 390, 265–278 (2001).
Jennewein, S., Long, R. M., Williams, R. M. & Croteau, R. Cytochrome p450 taxadiene 5alpha-hydroxylase, a mechanistically unusual monooxygenase catalyzing the first oxygenation step of taxol biosynthesis. Chem. Biol. 11, 379–387 (2004).
Dejong, J. M. et al. Genetic engineering of taxol biosynthetic genes in Saccharomyces cerevisiae. Biotechnol. Bioeng. 93, 212–224 (2006).
Li, H., Horiguchi, T., Croteau, R. & Williams, R. M. Studies on taxol biosynthesis: preparation of taxadiene-diol- and triol-derivatives by deoxygenation of taxusin. Tetrahedron 64, 6561–6567 (2008).
Jennewein, S., Rithner, C. D., Williams, R. M. & Croteau, R. B. Taxol biosynthesis: taxane 13 alpha-hydroxylase is a cytochrome P450-dependent monooxygenase. Proc. Natl Acad. Sci. USA 98, 13595–13600 (2001).
Kaspera, R. & Croteau, R. Cytochrome P450 oxygenases of Taxol biosynthesis. Phytochem. Rev. 5, 433–444 (2006).
Chau, M. & Croteau, R. Molecular cloning and characterization of a cytochrome P450 taxoid 2alpha-hydroxylase involved in Taxol biosynthesis. Arch. Biochem. Biophys. 427, 48–57 (2004).
Walker, K., Long, R. & Croteau, R. The final acylation step in taxol biosynthesis: cloning of the taxoid C13-side-chain N-benzoyltransferase from Taxus. Proc. Natl Acad. Sci. USA 99, 9166–9171 (2002).
Walker, K. & Croteau, R. Taxol biosynthetic genes. Phytochemistry 58, 1–7 (2001).
Walker, K. D., Klettke, K., Akiyama, T. & Croteau, R. Cloning, heterologous expression, and characterization of a phenylalanine aminomutase involved in Taxol biosynthesis. J. Biol. Chem. 279, 53947–53954 (2004).
Eisenreich, W., Menhard, B., Hylands, P. J., Zenk, M. H. & Bacher, A. Studies on the biosynthesis of taxol: the taxane carbon skeleton is not of mevalonoid origin. Proc. Natl Acad. Sci. USA 93, 6431–6436 (1996).
Ketchum, R. E. et al. Taxus metabolomics: methyl jasmonate preferentially induces production of taxoids oxygenated at C-13 in Taxus × media cell cultures. Phytochemistry 62, 901–909 (2003).
Sanchez-Muñoz, R. et al. A novel hydroxylation step in the taxane biosynthetic pathway: a new approach to paclitaxel production by synthetic biology. Front. Bioeng. Biotechnol. 8, 410 (2020).
Sabzehzari, M., Zeinali, M. & Naghavi, M. R. Alternative sources and metabolic engineering of Taxol: advances and future perspectives. Biotechnol. Adv. 43, 107569 (2020).
Ajikumar, P. K. et al. Isoprenoid pathway optimization for Taxol precursor overproduction in Escherichia coli. Science 330, 70–74 (2010).
Huang, Q., Roessner, C. A., Croteau, R. & Scott, A. I. Engineering Escherichia coli for the synthesis of taxadiene, a key intermediate in the biosynthesis of taxol. Bioorg. Med. Chem. 9, 2237–2242 (2001).
Engels, B., Dahm, P. & Jennewein, S. Metabolic engineering of taxadiene biosynthesis in yeast as a first step towards Taxol (Paclitaxel) production. Metab. Eng. 10, 201–206 (2008).
Besumbes, O. et al. Metabolic engineering of isoprenoid biosynthesis in Arabidopsis for the production of taxadiene, the first committed precursor of Taxol. Biotechnol. Bioeng. 88, 168–175 (2004).
Hasan, M. M. et al. Metabolic engineering of Nicotiana benthamiana for the increased production of taxadiene. Plant Cell Rep. 33, 895–904 (2014).
De La Peña, R. & Sattely, E. S. Rerouting plant terpene biosynthesis enables momilactone pathway elucidation. Nat. Chem. Biol. 17, 205–212 (2021).
Utomo, J. C., Chaves, F. C., Bauchart, P., Martin, V. J. J. & Ro, D. K. Developing a yeast platform strain for an enhanced taxadiene biosynthesis by CRISPR/Cas9. Metabolites 11, 147 (2021).
Rontein, D. et al. CYP725A4 from yew catalyzes complex structural rearrangement of taxa-4(5),11(12)-diene into the cyclic ether 5(12)-oxa-3(11)-cyclotaxane. J. Biol. Chem. 283, 6067–6075 (2008).
Edgar, S. et al. Mechanistic insights into taxadiene epoxidation by taxadiene-5α-hydroxylase. ACS Chem. Biol. 11, 460–469 (2016).
Li, C. et al. A cytochrome P450 enzyme catalyses oxetane ring formation in paclitaxel biosynthesis. Angew. Chem. Int. Ed. 63, e202407070 (2024).
Schoendorf, A., Rithner, C. D., Williams, R. M. & Croteau, R. B. Molecular cloning of a cytochrome P450 taxane 10β-hydroxylase cDNA from Taxus and functional expression in yeast. Proc. Natl Acad. Sci. USA 98, 1501–1506 (2001).
Chau, M., Walker, K., Long, R. & Croteau, R. Regioselectivity of taxoid-O-acetyltransferases: heterologous expression and characterization of a new taxadien-5alpha-ol-O-acetyltransferase. Arch. Biochem. Biophys. 430, 237–246 (2004).
Hampel, D., Mau, C. J. & Croteau, R. B. Taxol biosynthesis: identification and characterization of two acetyl CoA:taxoid-O-acetyl transferases that divert pathway flux away from Taxol production. Arch. Biochem. Biophys. 487, 91–97 (2009).
Li, B. J. et al. Improving 10-deacetylbaccatin III-10-β-O-acetyltransferase catalytic fitness for Taxol production. Nat. Commun. 8, 15544 (2017).
Ondari, M. E. & Walker, K. D. The taxol pathway 10-O-acetyltransferase shows regioselective promiscuity with the oxetane hydroxyl of 4-deacetyltaxanes. J. Am. Chem. Soc. 130, 17187–17194 (2008).
Lanier, E. R., Andersen, T. B. & Hamberger, B. Plant terpene specialized metabolism: complex networks or simple linear pathways? Plant J. 114, 1178–1201 (2023).
Xie, L., Gao, J. & Zhou, Y. J. Synthetic biology for Taxol biosynthesis and sustainable production. Trends Biotechnol. 42, 674–676 (2024).
Ramírez‐Estrada, K. et al. Transcript profiling of jasmonate‐elicited Taxus cells reveals a β‐phenylalanine‐CoA ligase. Plant Biotechnol. J. 14, 85–96 (2016).
Koetsier, M. J., Jekel, P. A., Wijma, H. J., Bovenberg, R. A. & Janssen, D. B. Aminoacyl-coenzyme A synthesis catalyzed by a CoA ligase from Penicillium chrysogenum. FEBS Lett. 585, 893–898 (2011).
Gou, Y., Jiang, X. & Lian, J. Intricate metabolic network for paclitaxel biosynthesis. BioDes. Res. https://doi.org/10.34133/bdr.0035 (2024).
Liu, X., Zhu, X., Cheng, J. & Jiang, H. A new era for paclitaxel biosynthesis is coming. Mol. Plant 17, 370–371 (2024).
Cope, E. A. Taxaceae: the genera and cultivated species. Bot. Rev. 64, 291–322 (1998).
Bundy, J. G., Davey, M. P. & Viant, M. R. Environmental metabolomics: a critical review and future perspectives. Metabolomics 5, 3–21 (2009).
Srividya, N. et al. Biochemical characterization of acyl activating enzymes for side chain moieties of Taxol and its analogs. J. Biol. Chem. 295, 4963–4973 (2020).
Nevarez, D. M., Mengistu, Y. A., Nawarathne, I. N. & Walker, K. D. An N-aroyltransferase of the BAHD superfamily has broad aroyl CoA specificity in vitro with analogues of N-dearoylpaclitaxel. J. Am. Chem. Soc. 131, 5994–6002 (2009).
Chen, X. Y., Wang, J. Q., Yang, Y., Li, J. & Chen, Z. S. Natural product as substrates of ABC transporters: a review. Recent Pat. Anticancer Drug Discov. 16, 222–238 (2021).
Wang, J. Q. et al. Multidrug resistance proteins (MRPs): structure, function and the overcoming of cancer multidrug resistance. Drug Resist. Updat. 54, 100743 (2021).
Syed, S. B. et al. Targeting P-glycoprotein: investigation of piperine analogs for overcoming drug resistance in cancer. Sci. Rep. 7, 7972 (2017).
Yu, C. et al. Integrated mass spectrometry imaging and single-cell transcriptome atlas strategies provide novel insights into taxoid biosynthesis and transport in Taxus mairei stems. Plant J. 115, 1243–1260 (2023).
Zheng, H. et al. Transcriptional regulatory network of high-value active ingredients in medicinal plants. Trends Plant Sci. 28, 429–446 (2023).
Ying, C. et al. miR5298b regulated taxol biosynthesis by acting on TcNPR3, resulting in an alleviation of the strong inhibition of the TcNPR3–TcTGA6 complex in Taxus chinensis. Int. J. Biol. Macromol. 248, 125909 (2023).
Zhang, M. et al. Transcriptome-wide identification and screening of WRKY factors involved in the regulation of taxol biosynthesis in Taxus chinensis. Sci. Rep. 8, 5197 (2018).
Zhang, K. et al. Transcriptome-wide analysis of AP2/ERF transcription factors involved in regulating Taxol biosynthesis in Taxus × media. Ind. Crops Prod. 171, 113972 (2021).
Zhan, X. et al. Mass spectrometry imaging and single-cell transcriptional profiling reveal the tissue-specific regulation of bioactive ingredient biosynthesis in Taxus leaves. Plant Commun. 4, 100630 (2023).
Chen, Y., Wang, Y., Liang, X., Zhang, Y. & Fernie, A. R. Mass spectrometric exploration of phytohormone profiles and signaling networks. Trends Plant Sci. https://doi.org/10.1016/j.tplants.2022.12.006 (2023).
Shen, S. et al. An Oryza-specific hydroxycinnamoyl tyramine gene cluster contributes to enhanced disease resistance. Sci. Bull. 66, 2369–2380 (2021).
Chakraborty, P. Gene cluster from plant to microbes: their role in genome architecture, organism’s development, specialized metabolism and drug discovery. Biochimie 193, 1–15 (2022).
Nützmann, H. W. & Osbourn, A. Gene clustering in plant specialized metabolism. Curr. Opin. Biotechnol. 26, 91–99 (2014).
Zhan, C. et al. Plant metabolic gene clusters in the multi-omics era. Trends Plant Sci. 27, 981–1001 (2022).
Boycheva, S., Daviet, L., Wolfender, J. L. & Fitzpatrick, T. B. The rise of operon-like gene clusters in plants. Trends Plant Sci. 19, 447–459 (2014).
Nützmann, H. W., Scazzocchio, C. & Osbourn, A. Metabolic gene clusters in eukaryotes. Annu. Rev. Genet. 52, 159–183 (2018).
Rhee, S. Y. & Mutwil, M. Towards revealing the functions of all genes in plants. Trends Plant Sci. 19, 212–221 (2014).
Purugganan, M. D. & Jackson, S. A. Advancing crop genomics from lab to field. Nat. Genet. 53, 595–601 (2021).
Stitt, M. Systems-integration of plant metabolism: means, motive and opportunity. Curr. Opin. Plant Biol. 16, 381–388 (2013).
Usadel, B. et al. Co-expression tools for plant biology: opportunities for hypothesis generation and caveats. Plant Cell Environ. 32, 1633–1651 (2009).
Patil, R. A., Kolewe, M. E., Normanly, J., Walker, E. L. & Roberts, S. C. Contribution of taxane biosynthetic pathway gene expression to observed variability in paclitaxel accumulation in Taxus suspension cultures. Biotechnol. J. 7, 418–427 (2012).
Li, S. T. et al. Transcriptional profile of Taxus chinensis cells in response to methyl jasmonate. BMC Genomics 13, 295 (2012).
Hong, B. et al. Biosynthesis of strychnine. Nature 607, 617–622 (2022).
Nett, R. S. et al. Plant carbonic anhydrase-like enzymes in neuroactive alkaloid biosynthesis. Nature 624, 182–191 (2023).
Liu, F., Fernie, A. R. & Zhang, Y. Plant gene co-expression defines the biosynthetic pathway of neuroactive alkaloids. Mol. Plant 17, 372–374 (2024).
Acknowledgements
F.L. and Y.Z. thank the National Natural Science Foundation of China (grant no. 32470416) for supporting this work.
Author information
Authors and Affiliations
Contributions
Y.Z. and A.R.F. jointly developed the logic and framework, conducted the literature review and led the overall writing process. F.L. summarized the main contents for each section, analysed the data and schematically summarized them into individual figures.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Plants thanks Zhihua Zhou and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Fig. 1.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Fernie, A.R., Liu, F. & Zhang, Y. Post-genomic illumination of paclitaxel biosynthesis. Nat. Plants 10, 1875–1885 (2024). https://doi.org/10.1038/s41477-024-01869-8
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41477-024-01869-8
This article is cited by
-
Engineered exosomes: a promising approach for overcoming challenges in pancreatic cancer therapy
Journal of Nanobiotechnology (2025)
-
Traditional Chinese medicine for sepsis: advancing from evidence to innovative drug discovery
Critical Care (2025)
-
Toward a Taxol-producing microbial cell factory
Nature Synthesis (2025)
-
Biochemical breakthrough paves way to reliable supply of anticancer drug
Nature (2025)
-
Synergistic effects of methyl-β-cyclodextrin and coronatine on biomass accumulation and taxane secretion in Corylus avellana L. cell suspension cultures
Plant Cell, Tissue and Organ Culture (PCTOC) (2025)