Abstract
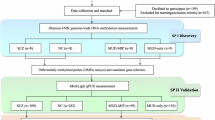
The dual-hit model of schizophrenic psychoses suggests that epigenetic alterations may contribute to the disease pathogenesis. Given the significant synaptic loss in patients with schizophrenia (SZ) during puberty, we investigated DNA-methylation patterns of key synaptic target molecules: dopamine transporter (DAT), dopamine receptor D2 (DRD2), microtubule-associated protein tau (MAPT), and postsynaptic density protein 95 (PSD95). Analyses were performed in both blood and cerebrospinal fluid (CSF) samples from patients with SZ (n = 36) and healthy controls (Co) (n = 23). Due to the minimal amount of cell-free DNA available in CSF, different extraction methods were evaluated to achieve the best possible recovery. Ultimately, an adapted ethanol-glycogen precipitation protocol combined with a subsequent bead-based fusion and DNA clean-up was applied. However, despite comparable DNA concentrations obtained from Co and SZ CSF samples, only very few sequences could be obtained from CSF samples of Co, so that results concerning CSF measurements are limited to patients with SZ. In DAT, methylation was significantly higher in the blood of Co compared to both the blood and CSF of patients with SZ. In PSD95, mean methylation levels were higher in the CSF than in the blood of patients with SZ, whereas no difference was detected in the blood between SZ and Co. For MAPT and DRD2, no significant differences in mean methylation rates were observed between groups. Low sequencing success in CSF from Co, despite comparable concentrations to SZ, might point to a higher degree of fragmentation. In SZ, longer DNA fragments may be replenished more frequently. Higher central methylation of PSD95 in patients with SZ, a key regulator of glutamatergic neurotransmission, may reduce gene transcription and thus support the glutamate hypothesis of SZ, which assumes impaired glutamate receptor function. Lower DAT methylation in SZ compared to Co (with similar central and peripheral levels) could indicate a higher availability of the transporter at the synapse in SZ, resulting in a higher clearance of dopamine. This could be a compensatory mechanism concerning the hypothesis of dopaminergic hyperactivity in SZ.
Similar content being viewed by others
Data availability
The data sets generated and analysed during the study are available from the corresponding author on reasonable request.
References
Bayer, T. A., Falkai, P. & Maier, W. Genetic and non-genetic vulnerability factors in schizophrenia: the basis of the “two hit hypothesis. J. Psychiatr. Res. 33, 543–548 (1999).
Barros, S. P. & Offenbacher, S. Epigenetics: connecting environment and genotype to phenotype and disease. J. Dent. Res 88, 400–408 (2009).
Kendler, K. S. A joint history of the nature of genetic variation and the nature of schizophrenia. Mol. Psychiatry 20, 77–83 (2015).
Ng, H. H. Bird A. DNA methylation and chromatin modification. Curr. Opin. Genet. Dev. 9, 158–163 (1999).
Goldberg, A. D., Allis, C. D. & Bernstein, E. Epigenetics: a landscape takes shape. Cell 128, 635–638 (2007).
Li, E., Bestor, T. H. & Jaenisch, R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69, 915–926 (1992).
Bennett, M. R. Synapse formation and regression in the cortex during adolescence and in schizophrenia. Med J. Aust. 190, S14–S16 (2009).
Sakka, L., Coll, G. & Chazal, J. Anatomy and physiology of cerebrospinal fluid. Eur. Ann. Otorhinolaryngol. Head. Neck Dis. 128, 309–316 (2011).
Sun, A. & Wang, J. Choroid plexus and drug removal mechanisms. AAPS J. 23, 61 (2021).
Brinker, T., Stopa, E., Morrison, J. & Klinge, P. A new look at cerebrospinal fluid circulation. Fluids Barriers CNS 11, 10 (2014).
Gilmore, J. H. & Bouldin, T. W. Analysis of ependymal abnormalities in subjects with schizophrenia, bipolar disorder, and depression. Schizophrenia Res. 57, 267–271 (2002).
Wojtkowska, M. et al. Quantification of circulating cell-free DNA in idiopathic Parkinson’s disease patients. Int. J. Mol. Sci. 25, 2818 (2024).
Southwood, D., Singh, S. & Chatterton, Z. Brain-derived cell-free DNA. Neural Regen. Res 17, 2213–2214 (2022).
Tripathy, A. et al. Liquid biopsy in pediatric brain tumors. Front Genet 13, 1114762 (2022).
Petty, A. et al. Enhanced Dopamine in Prodromal Schizophrenia (EDiPS): a new animal model of relevance to schizophrenia. NPJ Schizophr. 5, 6 (2019).
Ross, C. A., Margolis, R. L., Reading, S. A., Pletnikov, M. & Coyle, J. T. Neurobiology of schizophrenia. Neuron 52, 139–153 (2006
Purves-Tyson, T. D. et al. Putative presynaptic dopamine dysregulation in schizophrenia is supported by molecular evidence from post-mortem human midbrain. Transl. psychiatry 7, e1003 (2017).
Kruse, A. O. & Bustillo, J. R. Glutamatergic dysfunction in Schizophrenia. Transl. Psychiatry 12, 500 (2022).
Lerma, J. Roles and rules of kainate receptors in synaptic transmission. Nat. Rev. Neurosci. 4, 481–495 (2003).
Hu, W., MacDonald, M. L., Elswick, D. E. & Sweet, R. A. The glutamate hypothesis of schizophrenia: evidence from human brain tissue studies. Ann. N. Y. Acad. Sci. 1338, 38–57 (2015).
Coley, A. A. & Gao, W. J. PSD95: a synaptic protein implicated in schizophrenia or autism? Prog. Neuropsychopharmacol. Biol. Psychiatry 82, 187–194 (2018).
Kristiansen, L. V., Beneyto, M., Haroutunian, V. & Meador-Woodruff, J. H. Changes in NMDA receptor subunits and interacting PSD proteins in dorsolateral prefrontal and anterior cingulate cortex indicate abnormal regional expression in schizophrenia. Mol. Psychiatry 11, 737–747 (2006).
Jahn, K. et al. Differential methylation pattern of schizophrenia candidate genes in tetrahydrocannabinol-consuming treatment-resistant schizophrenic patients compared to non-consumer patients and healthy controls. Neuropsychobiology 80, 36–44 (2021).
Ruiz-Contreras, H. A. et al. Modulatory activity of the endocannabinoid system in the development and proliferation of cells in the CNS. Neurotox. Res 40, 1690–1706 (2022).
Demirel, O. F. et al. Total tau and phosphorylated tau protein serum levels in patients with schizophrenia compared with controls. Psychiatr. Q 88, 921–928 (2017
Miguel-Hidalgo, J. J., Dubey, P., Shao, Q., Stockmeier, C. & Rajkowska, G. Unchanged packing density but altered size of neurofilament immunoreactive neurons in the prefrontal cortex in schizophrenia and major depression. Schizophrenia Res. 76, 159–171 (2005).
Rovelet-Lecrux, A. & Campion, D. Copy number variations involving the microtubule-associated protein tau in human diseases. Biochem. Soc. Trans. 40, 672–676 (2012).
Deutsch, S. I., Rosse, R. B. & Lakshman, R. M. Dysregulation of tau phosphorylation is a hypothesized point of convergence in the pathogenesis of alzheimer’s disease, frontotemporal dementia and schizophrenia with therapeutic implications. Prog. Neuropsychopharmacol. Biol. Psychiatry 30, 1369–1380 (2006).
Lewin, J., Schmitt, A. O., Adorjan, P., Hildmann, T. & Piepenbrock, C. Quantitative DNA methylation analysis based on four-dye trace data from direct sequencing of PCR amplificates. Bioinformatics 20, 3005–3012 (2004).
Marques, D., Vaziri, N., Greenway, S. C. & Bousman, C. DNA methylation and histone modifications associated with antipsychotic treatment: a systematic review. Mol. Psychiatry 30, 296–309 (2025).
Rijcken, C. A., Monster, T. B., Brouwers, J. R. & de Jong-van den Berg, L. T. Chlorpromazine equivalents versus defined daily doses: how to compare antipsychotic drug doses? J. Clin. Psychopharmacol. 23, 657–659 (2003).
Schmauß, M. B. J. & Müller, W. E. Weiterbildungs-Curriculum psychopharmakologie/pharmaktherapie. Psychopharmakotherapie 26, 282–298 (2019).
Afflerbach, A. K. et al. Classification of brain tumors by nanopore sequencing of cell-free DNA from cerebrospinal fluid. Clin. Chem. 70, 250–260 (2024).
Ye, Z. et al. Cerebrospinal fluid liquid biopsy for detecting somatic mosaicism in brain. Brain Commun. 3, fcaa235 (2021).
Alcaide, M. et al. Evaluating the quantity, quality and size distribution of cell-free DNA by multiplex droplet digital PCR. Sci. Rep. 10, 12564 (2020).
Swarup, N. et al. Cell-free DNA: features and attributes shaping the next frontier in liquid biopsy. Mol. Diagn. Ther. 29, 277–290 (2025).
Choy, L. Y. L. et al. Single-molecule sequencing enables long cell-free DNA detection and direct methylation analysis for cancer patients. Clin. Chem. 68, 1151–1163 (2022).
Fan, Y., Abrahamsen, G., McGrath, J. J. & Mackay-Sim, A. Altered cell cycle dynamics in schizophrenia. Biol. psychiatry 71, 129–135 (2012).
Lubotzky, A. et al. Elevated brain-derived cell-free DNA among patients with first psychotic episode - a proof-of-concept study. eLife 11, e76391 (2022).
Coulter B. SPRI Bead to Sample Ratio for DNA Cleanup and Size Selection. https://www.beckman.es/reagents/genomic/cleanup-and-size-selection/pcr/bead-ratio2020.
Jahn, K. et al. Serotonin system-associated genetic and epigenetic changes in pedophilia and child sexual offending. J. Psychiatr. Res. 145, 60–69 (2022).
Long M. D., Smiraglia D. J., Campbell M. J. The genomic impact of DNA CpG methylation on gene expression; relationships in prostate cancer. Biomolecules 7, 15 (2017).
Wagner, J. R. et al. The relationship between DNA methylation, genetic and expression inter-individual variation in untransformed human fibroblasts. Genome Biol. 15, R37 (2014).
Jones, P. A. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 13, 484–492 (2012).
Javitt, D. C. Glutamate and schizophrenia: phencyclidine, N-methyl-D-aspartate receptors, and dopamine-glutamate interactions. Int. Rev. Neurobiol. 78, 69–108 (2007).
Guilmatre, A. et al. Recurrent rearrangements in synaptic and neurodevelopmental genes and shared biologic pathways in schizophrenia, autism, and mental retardation. Arch. Gen. Psychiatry 66, 947–956 (2009).
Acknowledgements
The authors would like to thank Andreas Niesel, Dept. of Neurology, Hannover Medical School, for expert technical assistance.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
Conceptualisation: K.J., S.B., and H.F. Methodology: K.J., O.R., S.S., and N.M. Software: K.J. and A.G. Formal Analysis: K.J. Investigation: K.J., A.G., F.K., and T.S. Resources: S.B., H.F., and T.S. Writing – original draft preparation: K.J. Writing – review and editing: A.G., O.R., S.S., N.M., S.B., H.F., F.K., and T.S. Visualisation: K.J. Supervision: S.B., H.F., and T.S. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jahn, K., Groh, A., Riemer, O. et al. Differential DNA-methylation of synaptic genes in CSF and blood in schizophrenia. Schizophr (2026). https://doi.org/10.1038/s41537-026-00738-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41537-026-00738-x