Abstract
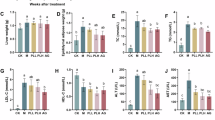
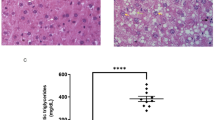
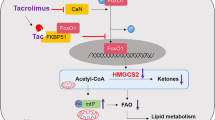
Hyperlipidemia is a leading global health challenge, limited by the safety liabilities of current pharmacotherapies. Here, we isolated a novel Limosilactobacillus reuteri strain, MacFasB02, from fecal samples of cynomolgus monkeys tolerant to chronic high-fat diet (HFD). This study aimed to systematically evaluate its probiotic properties and therapeutic potential against hyperlipidemia. In vitro, MacFasB02 exhibited robust growth, acid production, and tolerance to acidic and bile environments. In HFD-fed mice, 13-week MacFasB02 administration reduced weight gain, serum triglycerides, low-density lipoprotein cholesterol and total cholesterol, while ameliorating hepatic steatosis and inflammation, as well as restoring intestinal barrier integrity by enhanced villus architecture, goblet cell function, and tight junction proteins expression. Metagenomic analysis revealed gut microbiota remodeling. Transcriptomic profiling coupled with in vivo validation demonstrated upregulation of Apoa1 and Pltp in cholesterol metabolism. Untargeted metabolomics integrated with whole-genome sequencing and supernatant metabolite profiling identified adenosine as a key MacFasB02-derived metabolite in purine metabolism. Consistently, In vitro experiments showed that adenosine reduced lipid accumulation and inflammation in hepatocytes by regulating Apoa1 and Pltp to modulate cholesterol metabolism. Collectively, MacFasB02 exerts dual lipid-lowering and anti-inflammatory effects probably via adenosine-mediated modulation of cholesterol metabolism, promising potential as a live biopharmaceutical agent for hyperlipidemia.
Similar content being viewed by others
Data availability
The multi-omics data generated in this study have been deposited in the OMIX database (https://ngdc.cncb.ac.cn/omix) and GSA database (https://ngdc.cncb.ac.cn/gsub) at the China National Center for Bioinformation. OMIX013581: Non-targeted metabolomics of MacFasB02 culture medium Supernatant. Access link: https://ngdc.cncb.ac.cn/omix/release/OMIX013581. OMIX013583: Non-targeted metabolomics of mouse cecal contents. Access link: https://ngdc.cncb.ac.cn/omix/release/OMIX013583. CRA034905: Whole-genome sequencingof MacFasB02. Access link: https://ngdc.cncb.ac.cn/gsa/s/qwRF2z5T. CRA034902: Transcriptome sequencing of mouse liver samples. Access link: https://ngdc.cncb.ac.cn/gsa/s/XjRXx8S. CRA035096: Metagenomic Sequencing of mouse fecal samples. Access link: https://ngdc.cncb.ac.cn/gsa/s/22XFWxI1. CRA035456: Sanger sequencing identification of MacFasB02. Access link: https://ngdc.cncb.ac.cn/gsa/s/NBo8148Z.
References
Rasheed, M. & Chawla, G. Hyperlipidemia: a public health implication. ACADEMICIA: Int. Multidiscip. Res. J. 7, 84 (2017).
Su, X., Peng, H., Chen, X., Wu, X. & Wang, B. Hyperlipidemia and hypothyroidism. Clin. Chim. Acta 527, 61–70 (2022).
Gaggini, M., Gorini, F. & Vassalle, C. Lipids in Atherosclerosis: pathophysiology and the role of calculated lipid indices in assessing cardiovascular risk in patients with hyperlipidemia. Int. J. Mol. Sci. 24, https://doi.org/10.3390/ijms24010075 (2022).
Wei, F. et al. Nostoc sphaeroids Kütz ameliorates hyperlipidemia and maintains the intestinal barrier and gut microbiota composition of high-fat diet mice. Food Sci. Nutr. 8, 2348–2359 (2020).
Chaulin, A. Cardiotoxicity as a possible side effect of statins. Rev. Cardiovasc. Med. 24, 22 (2023).
Al-Shaer, M. H., Choueiri, N. E. & Suleiman, E. S. The pivotal role of cholesterol absorption inhibitors in the management of dyslipidemia. Lipids Health Dis. 3, 22 (2004).
Ferri, N. & Marodin, G. Emerging oral therapeutic strategies for inhibiting PCSK9. Atherosclerosis 59, 25–31 (2025).
Jiashuo, W. U., Fangqing, Z., Zhuangzhuang, L. I., Weiyi, J. & Yue, S. Integration strategy of network pharmacology in traditional chinese medicine: a narrative review. J. Tradit. Chin. Med. 42, 479–486 (2022).
Fan, Y. et al. Gut microbiota-targeted therapeutics for metabolic disorders: mechanistic insights into the synergy of probiotic-fermented herbal bioactives. Int. J. Mol. Sci. 26, https://doi.org/10.3390/ijms26125486 (2025).
Puttarat, N. et al. Beneficial effects of indigenous probiotics in high-cholesterol diet-induced hypercholesterolemic rats. Nutrients 15, https://doi.org/10.3390/nu15122710 (2023).
Cai, G., Wu, D., Li, X. & Lu, J. Levan from Bacillus amyloliquefaciens JN4 acts as a prebiotic for enhancing the intestinal adhesion capacity of Lactobacillus reuteri JN101. Int. J. Biol. Macromol. 146, 482–487 (2020).
Sun, Y. et al. Novel Lactobacillus reuteri HI120 Affects Lipid Metabolism in C57BL/6 Obese Mice. Front. Vet. Sci. 7, 560241 (2020).
Pakaew, K. et al. Lactobacillus reuteri TISTR 2736 alleviates type 2 diabetes in rats via the hepatic IRS1/PI3K/AKT signaling pathway by mitigating oxidative stress and inflammatory mediators. Eur. J. Nutr. 64, 27 (2024).
Mahemuti, N. et al. Association between systemic immunity-inflammation index and hyperlipidemia: a population-based study from the NHANES (2015-2020). Nutrients 15, https://doi.org/10.3390/nu15051177 (2023).
Liang, S. et al. Lactobacillus plantarum L11 and Lactobacillus reuteri LR: ameliorate obesity via AMPK Pathway. Nutrition 17, 4 (2025).
Jiang, J. et al. Lactobacillus reuteri A9 and lactobacillus mucosae A13 isolated from Chinese superlongevity people modulate lipid metabolism in a hypercholesterolemia rat model. FEMS Microbiol. Lett. 366, https://doi.org/10.1093/femsle/fnz254 (2019).
Yang, B. et al. Lactobacillus reuteri FYNLJ109L1 attenuating metabolic syndrome in mice via gut microbiota modulation and alleviating inflammation. Foods 10, https://doi.org/10.3390/foods10092081 (2021).
Gao, J. M. et al. Transplantation of gut microbiota from high-fat-diet-tolerant cynomolgus monkeys alleviates hyperlipidemia and hepatic steatosis in rats. Front. Microbiol. 13, 876043 (2022).
Wang, M. et al. Multilevel regulation of c-di-GMP biosynthesis by cAMP signaling increases Shigella sonnei fitness and pathogenicity in response to bile salts. Cell Rep. 44, 116306 (2025).
Makki, K., Deehan, E. C., Walter, J. & Bäckhed, F. The impact of dietary fiber on gut microbiota in host health and disease. Cell host microbe 23, 705–715 (2018).
Boranbayeva, G. et al. Probiotic consortium from poultry strains for supporting gut immunity against pathogens. Microb. Pathog. 204, 107584 (2025).
Panda, S. H., Goli, J. K., Das, S. & Mohanty, N. Production, optimization and probiotic characterization of potential lactic acid bacteria producing siderophores. AIMS Microbiol. 3, 88–107 (2017).
McConnell, E. L., Basit, A. W. & Murdan, S. Measurements of rat and mouse gastrointestinal pH, fluid and lymphoid tissue, and implications for in-vivo experiments. J. Pharm. Pharmacol. 60, 63–70 (2008).
Coman, M. M. et al. Probiotic characterization of Lactobacillus isolates from canine faeces. J. Appl. Microbiol. 126, 1245–1256 (2019).
Mohammadi, F. et al. Characterization of bacteriocin production in Lactobacillus spp. isolated from mother’s milk. Microb. Pathog. 118, 242–246 (2018).
Sobhani, S. et al. Body mass index, lipid profile, and hypertension contribute to prolonged QRS complex. Clin. Nutr. ESPEN 50, 231–237 (2022).
Wang, Y. et al. Gegen Qinlian Decoction Ameliorated DSS-induced colitis by attenuating inflammation, restoring intestinal mucosal barrier and modulating gut microbiota. J. Inflamm. Res. 18, 8065–8084 (2025).
Lee, M. et al. Lactobacillus reuteri NCIMB 30242 (LRC) inhibits cholesterol synthesis and stimulates cholesterol excretion in animal and cell models. J. Med. Food 26, 529–539 (2023).
Jia, S. et al. Changes of intestinal microbiome and its relationship with painful diabetic neuropathy in rats. BMC Microbiol. 25, 281 (2025).
Mu, Q., Tavella, V. J. & Luo, X. M. Role of lactobacillus reuteri in human health and diseases. Front. Microbiol. 9, 757 (2018).
Nagano, T., Higashimura, Y., Nakano, M., Nishiuchi, T. & Lelo, A. P. High-viscosity dietary fibers modulate gut microbiota and liver metabolism to prevent obesity in high-fat diet-fed mice. Int. J. Biol. Macromol. 298, 139962 (2025).
Chen, W. et al. Enhanced microbiota profiling in patients with quiescent Crohn’s disease through comparison with paired healthy first-degree relatives. Cell Rep. Med. 5, 101624 (2024).
Yang, Y. et al. Hippuric acid alleviates dextran sulfate sodium-induced colitis via suppressing inflammatory activity and modulating gut microbiota. Biochem. Biophys. Res. Commun. 710, 149879 (2024).
Lu, W. et al. Probiotic strains alleviated OVA-induced food allergy in mice by regulating the gut microbiota and improving the level of indoleacrylic acid in fecal samples. Food Funct. 13, 3704–3719 (2022).
Kather, H. Pathways of purine metabolism in human adipocytes. Further evidence against a role of adenosine as an endogenous regulator of human fat cell function. J. Biol. Chem. 265, 96–102 (1990).
Chaptal, M. C. et al. Low density lipoprotein cholesterol decreases the expression of adenosine A(2A) receptor and lipid rafts-protein flotillin-1: insights on cardiovascular risk of hypercholesterolemia. Cells 13, https://doi.org/10.3390/cells13060488 (2024).
Ingwersen, J. et al. Dual roles of the adenosine A2a receptor in autoimmune neuroinflammation. J. Neuroinflamm. 13, 48 (2016).
Wang, J. et al. Targeting the adenosine-mediated metabolic immune checkpoint with engineered probiotic for enhanced chemo-immunotherapy. Adv. Sci. 12, e2411813 (2025).
Min, H. K. et al. Increased hepatic synthesis and dysregulation of cholesterol metabolism is associated with the severity of nonalcoholic fatty liver disease. Cell Metab. 15, 665–674 (2012).
Pamir, N. et al. Proteomic analysis of HDL from inbred mouse strains implicates APOE associated with HDL in reduced cholesterol efflux capacity via the ABCA1 pathway. J. Lipid Res. 57, 246–257 (2016).
Gervois, P., Torra, I. P., Fruchart, J. C. & Staels, B. Regulation of lipid and lipoprotein metabolism by PPAR activators. Clin. Chem. Lab. Med. 38, 3–11 (2000).
Cai, H. et al. Dose-specific amelioration of caffeic acid phenethyl ester on high-fat diet-induced obesity based on intestinal FXR signaling and bile acid regulation. Food Biosci. 68, 106628 (2025).
Cui, D. et al. Cholesterol metabolism: molecular mechanisms, biological functions, diseases, and therapeutic targets. Mol. Biomed. 6, 72 (2025).
U.S. Food and Drug Administration. Redbook 2000: IV.B.1. General Guidelines for Designing and Conducting Toxicity Studies. Guidance for Industry and Other Stakeholders, https://www.fda.gov/regulatory-information/search-fda-guidance-documents/redbook-2000-ivb1-general-guidelines-designing-and-conducting-toxicity-studies (2007).
Organisation for Economic Co-operation and Development. Test No. 407: Repeated Dose 28-day Oral Toxicity Study in Rodents. OECD Guidelines for the Testing of Chemicals, https://doi.org/10.1787/9789264070684-en (2025).
Organisation for Economic Co-operation and Development. Test No. 408: Repeated Dose 90-day Oral Toxicity Study in Rodents. OECD Guidelines for the Testing of Chemicals, https://doi.org/10.1787/9789264070707-en (2025).
Zhang, C. et al. Lactobacillus reuteri J1 prevents obesity by altering the gut microbiota and regulating bile acid metabolism in obese mice. Food Funct. 13, 6688–6701 (2022).
Keresztény, T. et al. Isolation and characterization of lactic acid bacteria with probiotic attributes from different parts of the gastrointestinal tract of free-living wild boars in Hungary. Probiotics Antimicrob. Proteins 16, 1221–1239 (2024).
Karaseva, O. et al. Whole genome sequencing of the novel probiotic strain lactiplantibacillus plantarum FCa3L. Microorganisms 11, https://doi.org/10.3390/microorganisms11051234 (2023).
Gao, J. M. et al. Multiomics of parkinsonism cynomolgus monkeys highlights significance of metabolites in interaction between host and microbiota. NPJ Biofilms Microbiomes 10, 61 (2024).
Jin, Y., Teh, S. S., Lau, H. L. N. & Mah, S. H. In vivo toxicity assessment of refined red palm-pressed mesocarp olein in Sprague-Dawley Rats. J. Oleo Sci. 70, 1749–1759 (2021).
Liu, R. et al. Cytarabine chemotherapy induces meibomian gland dysfunction. Ocul. Surf. 34, 444–458 (2024).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 82471097 and 82200966), the Guangdong Key Laboratory of Nonhuman Primate Research (2020B121201006), Guangdong Provincial Science and Technology Leading Talent Project (210183503006). GDAS' Project of Science and Technology Development (2024GDASZH-2024010101).
Author information
Authors and Affiliations
Contributions
Y.J., H.J.A., J.H.R., and J.H.C. conceived and designed the study. Y.J., H.J.A., T.T.Z., J.M.G., X.L.Z., B.H.L., Y.Y.L., and X.J.Z. performed the experiments. Y.J., H.J.A., and J.J.L. analyzed the data. Y.J. wrote the original draft of the manuscript. Y.J., J.H.R., and J.H.C. contributed to writing, reviewing, and editing the manuscript. J.H.R. and J.H.C. supervised the research. J.H.R. and J.H.C. administered the project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Consent for publication
All authors approved the final manuscript and the submission to this journal.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Jin, Y., An, HJ., Zheng, TT. et al. Adenosine from high-fat-diet-tolerant monkey-derived Limsolactobacillus reuteri MacFasB02 modulates cholesterol metabolism to alleviate hyperlipidemia and inflammation. npj Sci Food (2026). https://doi.org/10.1038/s41538-026-00765-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41538-026-00765-z