Abstract
For brain–computer interfaces (BCIs), obtaining sufficient training data for algorithms that map neural signals onto actions can be difficult, expensive or even impossible. Here we report the development and use of a generative model—a model that synthesizes a virtually unlimited number of new data distributions from a learned data distribution—that learns mappings between hand kinematics and the associated neural spike trains. The generative spike-train synthesizer is trained on data from one recording session with a monkey performing a reaching task and can be rapidly adapted to new sessions or monkeys by using limited additional neural data. We show that the model can be adapted to synthesize new spike trains, accelerating the training and improving the generalization of BCI decoders. The approach is fully data-driven, and hence, applicable to applications of BCIs beyond motor control.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The raw and analysed datasets generated during the study are available on Github at https://github.com/shixianwen/Rapid-transfer-of-brain-machine-interfaces-to-new-neuronal-ensembles-or-participants.
Code availability
The codes used in this study are available on Github at https://github.com/shixianwen/Rapid-transfer-of-brain-machine-interfaces-to-new-neuronal-ensembles-or-participants.
References
Wessberg, J. et al. Real-time prediction of hand trajectory by ensembles of cortical neurons in primates. Nature https://doi.org/10.1038/35042582 (2000).
Velliste, M., Perel, S., Spalding, M. C., Whitford, A. S. & Schwartz, A. B. Cortical control of a prosthetic arm for self-feeding. Nature https://doi.org/10.1038/nature06996 (2008).
Taylor, D. M., Tillery, S. I. H. & Schwartz, A. B. Direct cortical control of 3D neuroprosthetic devices. Science https://doi.org/10.1126/science.1070291 (2002).
Carmena, J. M. et al. Learning to control a brain–machine interface for reaching and grasping by primates. PLoS Biol. https://doi.org/10.1371/journal.pbio.0000042 (2003).
Li, Z. et al. Unscented Kalman filter for brain–machine interfaces. PLoS ONE https://doi.org/10.1371/journal.pone.0006243 (2009).
Wu, W. et al. Neural decoding of cursor motion using a Kalman filter. In Proc. Advances in Neural Information Processing Systems 15 (eds Becker, S., Thrun, S. & Obermayer, K.) 133-140 (MIT Press, 2003).
Brockwell, A. E. Recursive Bayesian decoding of motor cortical signals by particle filtering. J. Neurophysiol. https://doi.org/10.1152/jn.00438.2003 (2004).
Gao, Y., Black, M. J., Bienenstock, E., Wu, W. & Donoghue, J. P. A quantitative comparison of linear and non-linear models of motor cortical activity for the encoding and decoding of arm motions. In First International IEEE EMBS Conference on Neural Engineering (IEEE, 2003); https://doi.org/10.1109/CNE.2003.1196789
Eden, U. T., Frank, L. M., Barbieri, R., Solo, V. & Brown, E. N. Dynamic analysis of neural encoding by point process adaptive filtering. Neural Comput. https://doi.org/10.1162/089976604773135069 (2004).
Eden, U. T. Point process adaptive filters for neural data analysis: theory and applications. In Proc. IEEE Conference on Decision and Control (IEEE, 2007); https://doi.org/10.1109/CDC.2007.4434708
Hochreiter, S. & Schmidhuber, J. Long short-term memory. Neural Comput. https://doi.org/10.1162/neco.1997.9.8.1735 (1997).
Glaser, J. I. et al. Machine learning for neural decoding. Eneuro https://doi.org/10.1523/ENEURO.0506-19.2020 (2020).
Moeendarbary, E. et al. The soft mechanical signature of glial scars in the central nervous system. Nat. Commun. https://doi.org/10.1038/ncomms14787 (2017).
Kozai, T. D. Y., Jaquins-Gerstl, A. S., Vazquez, A. L., Michael, A. C. & Cui, X. T. Brain tissue responses to neural implants impact signal sensitivity and intervention strategies. ACS Chem. Neurosci. https://doi.org/10.1021/cn500256e (2015).
Duffau, H. Brain plasticity and reorganization before, during, and after glioma resection. Glioblastoma https://doi.org/10.1016/B978-0-323-47660-7.00018-5 (2016).
Tkach, D., Reimer, J. & Hatsopoulos, N. G. Observation-based learning for brain–machine interfaces. Curr. Opin. Neurobiol. 18, 589–594 (2008).
Gallego, J. A., Perich, M. G., Chowdhury, R. H., Solla, S. A. & Miller, L. E. Long-term stability of cortical population dynamics underlying consistent behavior. Nat. Neurosci. 23, 260–270 (2020).
Gao, P. et al. A theory of multineuronal dimensionality, dynamics and measurement. Preprint at bioRxiv https://doi.org/10.1101/214262 (2017).
Pandarinath, C. et al. Inferring single-trial neural population dynamics using sequential auto-encoders. Nat. Methods https://doi.org/10.1038/s41592-018-0109-9 (2018).
Farshchian, A. et al. Adversarial domain adaptation for stable brain-machine interfaces. Seventh International Conference on Learning Representations Paper 1437 (ICLR, 2019).
Sussillo, D., Stavisky, S. D., Kao, J. C., Ryu, S. I. & Shenoy, K. V. Making brain–machine interfaces robust to future neural variability. Nat. Commun. https://doi.org/10.1038/ncomms13749 (2016).
Degenhart, A. D. et al. Stabilization of a brain–computer interface via the alignment of low-dimensional spaces of neural activity. Nat. Biomed. Eng. 4, 672–685 (2020).
Gerstner, W., Kistler, W. M., Naud, R. & Paninski, L. Neuronal Dynamics: From Single Neurons to Networks and Models of Cognition (Cambridge Univ. Press, 2014); https://doi.org/10.1017/CBO9781107447615
Goodfellow, I. et al. Generative adversarial nets. in Advances in Neural Information Processing Systems 27 (2014); https://papers.nips.cc/paper/2014/hash/5ca3e9b122f61f8f06494c97b1afccf3-Abstract.html
Wei, L., Hu, L., Kim, V., Yumer, E. & Li, H. Real-time hair rendering using sequential adversarial networks. In Computer Vision – ECCV 2018. ECCV 2018. Lecture Notes in Computer Science Vol. 11208 (eds Ferrari V., Hebert M., Sminchisescu C. & Weiss Y.) 105–122 (Springer, 2018); https://doi.org/10.1007/978-3-030-01225-0_7
Jetchev, N. & Bergmann, U. The conditional analogy GAN: swapping fashion articles on people images. In 2017 IEEE International Conference on Computer Vision Workshops, ICCVW 2017 (IEEE, 2018); https://doi.org/10.1109/ICCVW.2017.269
Chen, Xi. et al. InfoGAN: interpretable representation learning by information maximizing generative adversarial nets. Preprint at https://arxiv.org/abs/1606.03657 (2016).
Mirza, M. & Osindero, S Conditional generative adversarial nets. Preprint at https://arxiv.org/abs/1411.1784 (2014).
Odena, A, Olah, C. & Shlens, J. Conditional image synthesis with auxiliary classifier GANs. Proc. 34th International Conference on Machine Learning (eds Precup, D. & Teh, Y. W.) 2642–2651 (PMLR, 2017).
Ho, D., Liang, D., Stoica, I., Abbeel, P. & Chen, X. Population based augmentation: efficient learning of augmentation policy schedules. Proc. 36th International Conference on Machine Learning (eds Chaudhuri, K. & Salakhutdinov, R.) 2731–2741 (PMLR, 2019).
Dai, W., Yang, Q., Xue, G.-R. & Yu, Y. Boosting for transfer learning. In Proceedings of the 24th International Conference on Machine learning - ICML ’07 (ACM, 2007); https://doi.org/10.1145/1273496.1273521
Arnold, A., Nallapati, R. & Cohen, W. W. A comparative study of methods for transductive transfer learning. In Proceedings - IEEE International Conference on Data Mining, ICDM (IEEE, 2007); https://doi.org/10.1109/ICDMW.2007.109
Lin, Min, Qiang Chen, and Shuicheng Yan. “Network in network.” arXiv preprint arXiv:1312.4400 (2013).
Tchumatchenko, T., Geisel, T., Volgushev, M. & Wolf, F. Spike correlations - what can they tell about synchrony? Front. Neurosci. https://doi.org/10.3389/fnins.2011.00068 (2011).
Dyer, E. L. et al. A cryptography-based approach for movement decoding. Nat. Biomed. Eng. https://doi.org/10.1038/s41551-017-0169-7 (2017).
Ijspeert, A. J., Nakanishi, J., Hoffmann, H., Pastor, P. & Schaal, S. Dynamical movement primitives: learning attractor models for motor behaviors. Neural Comput. https://doi.org/10.1162/NECO_a_00393 (2013).
Schaal, S. “Dynamic movement primitives-a framework for motor control in humans and humanoid robotics.” Adaptive motion of animals and machines. Springer, Tokyo, 2006. 261–280.
Poggio, T. & Bizzi, E. Generalization in vision and motor control. Nature https://doi.org/10.1038/nature03014 (2004).
Nuyujukian, P. et al. Performance sustaining intracortical neural prostheses. J. Neural Eng. https://doi.org/10.1088/1741-2560/11/6/066003 (2014).
Thoroughman, K. A. & Shadmehr, R. Learning of action through adaptive combination of motor primitives. Nature https://doi.org/10.1038/35037588 (2000).
Stroud, J. P., Porter, M. A., Hennequin, G. & Vogels, T. P. Motor primitives in space and time via targeted gain modulation in cortical networks. Nat. Neurosci. https://doi.org/10.1038/s41593-018-0276-0 (2018).
Shankar, T., Pinto, L., Tulsiani, S. & Gupta, A. Discovering motor programs by recomposing demonstrations. Conference paper 1246 in International Conference on Learning Representations (ICLR, 2020).
Costa, R. M., Ganguly, K., Costa, R. M. & Carmena, J. M. Emergence of coordinated neural dynamics underlies neuroprosthetic learning and skillful control. Neuron https://doi.org/10.1016/j.neuron.2017.01.016 (2017).
Golub, M. D. et al. Learning by neural reassociation. Nat. Neurosci. https://doi.org/10.1038/s41593-018-0095-3 (2018).
Sadtler, P. T. et al. Neural constraints on learning. Nature https://doi.org/10.1038/nature13665 (2014).
Gold, J. I. & Shadlen, M. N. The neural basis of decision making. Annu. Rev. Neurosci. https://doi.org/10.1146/annurev.neuro.29.051605.113038 (2007).
Felsen, G. & Dan, Y. A natural approach to studying vision. Nat. Neurosci. https://doi.org/10.1038/nn1608 (2005).
Paninski, L., Pillow, J. & Lewi, J. Statistical models for neural encoding, decoding, and optimal stimulus design. Prog. Brain Res. https://doi.org/10.1016/S0079-6123(06)65031-0 (2007).
Paninski, L. Superlinear population encoding of dynamic hand trajectory in primary motor cortex. J. Neurosci. https://doi.org/10.1523/JNEUROSCI.0919-04.2004 (2004).
Srivastava, N., Hinton, G., Krizhevsky, A., Sutskever, I. & Salakhutdinov, R. Dropout: a simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 15, 1929–1958 (2014).
Senior, A. et al. An empirical study of learning rates in deep neural networks for speech recognition. In 2013 IEEE International Conference on Acoustics, Speech and Signal Processing. 6724–6728 (IEEE, 2013).
Schuster, M. & Paliwal, K. K. Bidirectional recurrent neural networks. IEEE Trans. Signal Process. 45, 2673–2681 (1997).
le Cun, Y. A theoretical framework for back-propagation. In Proc. 1988 Connectionist Models Summer School Vol. 1. (eds Touretzky, D., Hinton, G. & Sejnowski, T.) 21–28 (CMU, 1988).
LeCun, Y. A., Bottou, L., Orr, G. B. & Müller, K. R. In Neural Networks: Tricks of the Trade 2nd edn (eds Montavon, G., Orr, G. B. & Müller, K. R.) 9–48 (Springer, 2012); https://doi.org/10.1007/978-3-642-35289-8_3
Acknowledgements
This work was supported by the National Science Foundation (grant no. CCF-1317433), C-BRIC (one of six centers in JUMP, a Semiconductor Research Corporation (SRC) program sponsored by DARPA), the Intel Corporation and the National Institutes of Health (grant nos. NIH NINDS T32 HD07418, F31 NS092356, NS053603 and NS074044). We affirm that the views expressed herein are solely our own and do not represent the views of the US government or any agency thereof.
Author information
Authors and Affiliations
Contributions
M.G.P. and L.E.M. conducted the experiments. S.W., A.Y., T.F. and L.I. analysed the results. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Biomedical Engineering thanks Jonathan Viventi and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 General Framework.
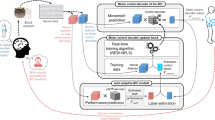
Step 1: training a neural spike synthesizer on the neural data from session one of Monkey one to learn a direct mapping from kinematics to spike trains and to capture the embedded neural attributes. Step 2: freezing the generator that captures the embedded neural attributes and fine-tuning the readout modules for different sessions or subjects to allow variations in neural attributes, using the neural data from session two of Monkey one or the neural data from session one of Monkey two. Then, synthesizing a large amount of spike trains that are suitable for another session or subject. Step 3: training a BCI decoder for another session or subject using the same small amount of real neural data used for fine-tuning (in step 2) and a large amount of synthesized spike trains (in step 2). Step 4: testing the same BCI decoder on an independent test set from another session or subject.
Extended Data Fig. 2 Cross-session decoding.
The GAN-augmentation, mutation-augmentation, stretch-augmentation, real-concatenation and real-only methods are shown in red, purple, orange, blue and green curves with an error bar in 5-fold cross-validation. The horizontal axis is the number of minutes of neural data from the session two of Monkey C used. The vertical axis is correlation coefficient between the decoded kinematics and real kinematics on an independent test set from the session two of Monkey C (mean +/ - S.D., n = 5 folds). Synthesized spike trains that capture the neural attributes accelerate the training of a BCI decoder for the cross-session decoding.
Extended Data Fig. 3 Cross-subject decoding.
The GAN-augmentation, mutation-augmentation, stretch-augmentation, real-concatenation and real-only methods are shown in red, purple, orange, blue and green curves with an error bar in 5-fold cross-validation. The horizontal axis is the number of minutes of neural data from Monkey M used. The vertical axis is the correlation coefficient between the decoded kinematics and real kinematics on an independent test set from the Monkey M (mean + / - S.D., n = 5 folds). When the neural data from another subject is limited, synthesized spike trains that capture the neural attributes improve the cross-subject decoding performance on acceleration. Even with ample neural data for both subjects, the neural attributes learned from one subject can transfer some useful knowledge that improves the best achievable decoding performance on the acceleration of another subject.
Extended Data Fig. 4
Detailed structure of the CC-LSTM-GAN.
Supplementary information
Supplementary Information
Supplementary figures, discussion and references.
Rights and permissions
About this article
Cite this article
Wen, S., Yin, A., Furlanello, T. et al. Rapid adaptation of brain–computer interfaces to new neuronal ensembles or participants via generative modelling. Nat. Biomed. Eng 7, 546–558 (2023). https://doi.org/10.1038/s41551-021-00811-z
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41551-021-00811-z
This article is cited by
-
KA-AttLSTMnet: a Kolmogorov-Arnold attentional architecture for egocentric navigation prediction from hippocampal CA1 spikes
Applied Intelligence (2026)
-
A memristor-based adaptive neuromorphic decoder for brain–computer interfaces
Nature Electronics (2025)
-
Generative motor imagery dynamic networks: EEG-controlled grasping via individualized model training
Cognitive Neurodynamics (2025)
-
Rethink the motor cortical control via the experiment-analysis-model flywheel: an overview
Med-X (2025)
-
Recent applications of EEG-based brain-computer-interface in the medical field
Military Medical Research (2025)