Abstract
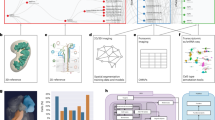
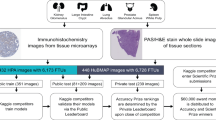
The Human BioMolecular Atlas Program (HuBMAP) aims to create a multi-scale spatial atlas of the healthy human body at single-cell resolution by applying advanced technologies and disseminating resources to the community. As the HuBMAP moves past its first phase, creating ontologies, protocols and pipelines, this Perspective introduces the production phase: the generation of reference spatial maps of functional tissue units across many organs from diverse populations and the creation of mapping tools and infrastructure to advance biomedical research.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Code availability
The HuBMAP Consortium GitHub at https://github.com/hubmapconsortium has 120 repositories that support data ingest, analysis, visualization and search plus HRA construction and usage. Documentation is available at https://software.docs.hubmapconsortium.org/apis.html.
Change history
01 March 2024
A Correction to this paper has been published: https://doi.org/10.1038/s41556-024-01384-0
References
Snyder, M. P. et al. The human body at cellular resolution: the NIH Human Biomolecular Atlas Program. Nature 574, 187–192 (2019).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021).
Börner, K. et al. Anatomical structures, cell types and biomarkers of the Human Reference Atlas. Nat. Cell Biol. 23, 1117–1128 (2021).
Hickey, J. W. et al. Spatial mapping of protein composition and tissue organization: a primer for multiplexed antibody-based imaging. Nat. Methods 19, 284–295 (2022).
Quardokus, E. M. et al. Organ Mapping Antibody Panels: a community resource for standardized multiplexed tissue imaging. Nat. Methods https://doi.org/10.1038/s41592-023-01846-7 (2023).
Neumann, E. K. et al. A multiscale lipid and cellular atlas of the human kidney. Preprint at bioRxiv https://doi.org/10.1101/2022.04.07.487155 (2022).
Ghose, S. et al. Human digital twin: automated cell type distance computation and 3D atlas construction in multiplexed skin biopsies. Preprint at bioRxiv https://doi.org/10.1101/2022.03.30.486438 (2022).
Canela, V. H. et al. A spatially anchored transcriptomic atlas of the human kidney papilla identifies significant immune injury and matrix remodeling in patients with stone disease. Nat. Commun. https://doi.org/10.1038/s41467-023-38975-8 (2023).
Hickey, J. et al. High resolution single cell maps reveals distinct cell organization and function across different regions of the human intestine. Nature https://doi.org/10.1038/s41586-023-05915-x (2023).
Greenbaum, S. et al. Spatiotemporal coordination at the maternal–fetal interface promotes trophoblast invasion and vascular remodeling in the first half of human pregnancy. Nature https://doi.org/10.1038/s41586-023-06298-9 (2023).
Blue, B. L. et al. An atlas of healthy and injured cell states and niches in the human kidney. Nature https://doi.org/10.1038/s41586-023-05769-3 (2023).
Eng, C.-H. L. et al. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH. Nature 568, 235–239 (2019).
Rodriques, S. G. et al. Slide-seq: a scalable technology for measuring genome-wide expression at high spatial resolution. Science 363, 1463–1467 (2019).
Guilliams, M. et al. Spatial proteogenomics reveals distinct and evolutionarily conserved hepatic macrophage niches. Cell 185, 379–396.e38 (2022).
Stelzer, E. H. K. Light-sheet fluorescence microscopy for quantitative biology. Nat. Methods 12, 23–26 (2015).
Black, S. et al. CODEX multiplexed tissue imaging with DNA-conjugated antibodies. Nat. Protoc. 16, 3802–3835 (2021).
Neumann, E. K. et al. Protocol for multimodal analysis of human kidney tissue by imaging mass spectrometry and CODEX multiplexed immunofluorescence. STAR Protoc. 2, 100747 (2021).
Spraggins, J. M. et al. High-performance molecular imaging with MALDI trapped ion-mobility time-of-flight (timsTOF) mass spectrometry. Anal. Chem. 91, 14552–14560 (2019).
Börner, K. et al. Tissue registration and exploration user interfaces in support of a human reference atlas. Commun. Biol. 5, 1369 (2022).
Rozenblatt-Rosen, O. et al. The Human Tumor Atlas Network: charting tumor transitions across space and time at single-cell resolution. Cell 181, 236–249 (2020).
Rozenblatt-Rosen, O., Stubbington, M. J. T., Regev, A. & Teichmann, S. A. The Human Cell Atlas: from vision to reality. Nature 550, 451–453 (2017).
Mungall, C. J., Torniai, C., Gkoutos, G. V., Lewis, S. E. & Haendel, M. A. Uberon, an integrative multi-species anatomy ontology. Genome Biol. 13, R5 (2012).
Rosse, C. & Mejino, J. L. V. A reference ontology for biomedical informatics: the Foundational Model of Anatomy. J. Biomed. Inform. 36, 478–500 (2003).
Golbreich, C., Grosjean, J. & Darmoni, S. J. The Foundational Model of Anatomy in OWL 2 and its use. Artif. Intell. Med. 57, 119–132 (2013).
Meehan, T. F. et al. Logical development of the cell ontology. BMC Bioinform. 12, 6 (2011).
Eraslan, G. et al. Single-nucleus cross-tissue molecular reference maps toward understanding disease gene function. Science 376, eabl4290 (2022).
de Boer, I. H. et al. Rationale and design of the Kidney Precision Medicine Project. Kidney Int. 99, 496–510 (2021).
Manz, T. et al. Viv: multiscale visualization of high-resolution multiplexed bioimaging data on the web. Nat. Methods 19, 515–516 (2022).
Lee, P. J. et al. NIH SenNet Consortium to map senescent cells throughout the human lifespan to understand physiological health. Nat. Aging 2, 1090–1100 (2022).
Keren, L. et al. MIBI-TOF: a multiplexed imaging platform relates cellular phenotypes and tissue structure. Sci. Adv. 5, eaax5851 (2019).
He, S. et al. High-plex imaging of RNA and proteins at subcellular resolution in fixed tissue by spatial molecular imaging. Nat. Biotechnol. 40, 1794–1806 (2022).
Tian, H. et al. Successive high-resolution (H2O)n-GCIB and C60-SIMS imaging integrates multi-omics in different cell types in breast cancer tissue. Anal. Chem. 93, 8143–8151 (2021).
Xu, T.-Y., Calandrelli, R., Lin, P., Zhu, D. & Zhong, S. HiFi-Slide spatial RNA-sequencing. Protocol.io https://doi.org/10.17504/protocols.io.3byl4by18vo5/v1 (2022).
Fung, A. A. & Shi, L. Mammalian cell and tissue imaging using Raman and coherent Raman microscopy. Wiley Interdiscip. Rev. Syst. Biol. Med. 12, e1501 (2020).
Zhang, W. et al. Multi-molecular hyperspectral PRM-SRS imaging. Preprint at bioRxiv https://doi.org/10.1101/2022.07.25.501472 (2022).
Balaratnasingam, C. et al. Histologic and optical coherence tomographic correlates in drusenoid pigment epithelium detachment in age-related macular degeneration. Ophthalmology 124, 644–656 (2017).
Spaide, R. F. & Curcio, C. A. Anatomical correlates to the bands seen in the outer retina by optical coherence tomography: literature review and model. Retina 31, 1609 (2011).
Zhu, Y. et al. Nanodroplet processing platform for deep and quantitative proteome profiling of 10–100 mammalian cells. Nat. Commun. 9, 882 (2018).
Ardini-Poleske, M. E. et al. LungMAP: the molecular atlas of lung development program. Am. J. Physiol. Lung Cell. Mol. Physiol. 313, L733–L740 (2017).
Uhlén, M. et al. Tissue-based map of the human proteome. Science 347, 1260419 (2015).
Lonsdale, J. et al. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 45, 580–585 (2013).
Brbić, M. et al. Annotation of spatially resolved single-cell data with STELLAR. Nat. Methods 19, 1411–1418 (2022).
Govind, D. et al. PodoSighter: A cloud-based tool for label-free podocyte detection in kidney whole-slide images. J. Am. Soc. Nephrol. 32, 2795–2813 (2021).
Lutnick, B. et al. A user-friendly tool for cloud-based whole slide image segmentation with examples from renal histopathology. Commun. Med. 2, 105–109 (2022).
Van de Plas, R., Yang, J., Spraggins, J. & Caprioli, R. M. Image fusion of mass spectrometry and microscopy: a multimodality paradigm for molecular tissue mapping. Nat. Methods 12, 366–372 (2015).
Tideman, L. E. M. et al. Automated biomarker candidate discovery in imaging mass spectrometry data through spatially localized Shapley additive explanations. Anal. Chim. Acta 1177, 338522 (2021).
Weber, G. M., Ju, Y. & Börner, K. Considerations for using the vasculature as a coordinate system to map all the cells in the human body. Front. Cardiovasc. Med. 7, 29 (2020).
Melani, R. D. et al. The Blood Proteoform Atlas: a reference map of proteoforms in human hematopoietic cells. Science 375, 411–418 (2022).
Mackenzie, N. J. et al. Modelling the tumor immune microenvironment for precision immunotherapy. Clin. Transl. Immunol. 11, e1400 (2022).
Becker, W. R. et al. Single-cell analyses define a continuum of cell state and composition changes in the malignant transformation of polyps to colorectal cancer. Nat. Genet. 54, 985–995 (2022).
Wallace, D. C. Mitochondrial DNA variation in human radiation and disease. Cell 163, 33–38 (2015).
Wallace, D. C. Mitochondrial genetic medicine. Nat. Genet. 50, 1642–1649 (2018).
Acknowledgements
We thank L. M. McGuire, SciStories, F. Goncalves, H. Schlehlein and V. A. Deshpande for their efforts in designing and creating graphics. We thank A. Honkala for assistance with manuscript formatting. We thank Z. Galis from the National Heart, Lung, and Blood Institute for many useful comments. Support for title page creation and format was provided by AuthorArranger, a tool developed at the National Cancer Institute. The authors gratefully acknowledge NIH HuBMAP grants U54HL165442 and U01HL166058 (L.P.); U54DK134301 and OT2OD033753 (S.J.); U54EY032442, U54DK120058 and U54DK134302 (J.M.S.); OT2OD033756 and OT2OD026671 (K.B.); UH3CA246635 (N.L.K.); UG3CA256967 (H.L.); U54HG010426 (M.P.S.); UH3CA246633 (M.A.); U54HD104393 (L.C.L. and P.R.); U54DK127823 (E.S.N., J.P.C. and W.-J.Q.); OT2OD033758 (N.G.); UH3CA246594 (F.G.); U54EY032442 and U54DK134302 (J.P.G.); U54HL145611 (S.L. and Y.L.); U01HG012680 (A.N.); U54DK120058, U54EY032442 and U54DK134302 (B.R.S.); UG3CA256959 (M.Z.); and U54HL165440 (I.S.V.). The authors are also supported by these grants: 2U01DK114933, P50DK133943 and U24DK135157 (S.J.); U54HL145608 and U54HL165443 (J.S.H.); U24CA268108 and U2CDK114886 (K.B.); Department of Defense W81XWH-22-1-0058 and Additional Ventures (L.P.). J.W.H. was supported by an NIH T32 Fellowship (T32CA196585) and an American Cancer Society: Roaring Fork Valley Postdoctoral Fellowship (PF-20-032-01-CSM). This work was supported, in part, by the Intramural Research Program of the NIH, National Institute of Allergy and Infectious Diseases (NIAID) and National Cancer Institute (NCI) (A.J.R.).
Author information
Authors and Affiliations
Consortia
Contributions
S.J., L.P., J.M.S., K.B. and M.P.S. conceived the idea and wrote the first draft of the manuscript and revisions. M.A., J.P.C., N.G., F.G., J.P.G., J.S.H., J.W.H., N.L.K., L.C.L., S.L., Y.L., H.L., A.N., E.S.N., W.-J.Q., A.R., P.R., B.R.S., R.V., I.S.V. and M.Z. contributed to manuscript revision.
Corresponding authors
Ethics declarations
Competing interests
The authors declare the following competing interests. F.G. is an employee of GE Research. B.R.S. is an inventor on patents and patent applications involving small-molecule-drug discovery and the 3F3 anti-Ferroptotic Membrane (3F3-FMA) antibody; co-founded and serves as a consultant to Inzen Therapeutics, Nevrox, Exarta Therapeutics and ProJenX; and serves as a consultant to Weatherwax Biotechnologies and Akin Gump Strauss Hauer and Feld. M.P.S. is a co-founder and an advisory board member of Personalis, Qbio, January AI, Mirvie, Filtricine, Fodsel, Lollo and Protos. I.S.V. consults for Guidepoint Global, Cowen, Mosaic and NextRNA. N.G. is a co-founder and an equity owner of Datavisyn. H.L. is a co-founder and an equity owner of ExoMira Medicine. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Cell Biology thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jain, S., Pei, L., Spraggins, J.M. et al. Advances and prospects for the Human BioMolecular Atlas Program (HuBMAP). Nat Cell Biol 25, 1089–1100 (2023). https://doi.org/10.1038/s41556-023-01194-w
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41556-023-01194-w
This article is cited by
-
Spatial top-down proteomics for the functional characterization of human kidney
Clinical Proteomics (2025)
-
The Human Cell Atlas from a cell census to a unified foundation model
Nature (2025)
-
The spatial and temporal activation of macrophages during fibrosis
Nature Reviews Immunology (2025)
-
Volumetric imaging using a distributed molecular network
Nature Biotechnology (2025)
-
Three dimensional multiscalar neurovascular nephron connectivity map of the human kidney across the lifespan
Nature Communications (2025)