Abstract
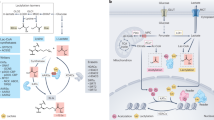
Given its various roles in cellular functions, lactate is no longer considered a waste product of metabolism and lactate sensing is a pivotal step in the transduction of lactate signals. Lysine lactylation is a recently identified post-translational modification that serves as an intracellular mechanism of lactate sensing and transfer. Although acetyltransferases such as p300 exhibit general acyl transfer activity, no bona fide lactyltransferases have been identified. Recently, the protein synthesis machinery, alanyl-tRNA synthetase 1 (AARS1), AARS2 and their Escherichia coli orthologue AlaRS, have been shown to be able to sense lactate and mediate lactyl transfer and are thus considered pan-lactyltransferases. Here we highlight the mechanisms and functions of these lactyltransferases and discuss potential strategies that could be exploited for the treatment of human diseases.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Ippolito, L., Morandi, A., Giannoni, E. & Chiarugi, P. Lactate: a metabolic driver in the tumour landscape. Trends Biochem. Sci. 44, 153–166 (2019).
Jansen, T. C., van Bommel, J. & Bakker, J. Blood lactate monitoring in critically ill patients: a systematic health technology assessment. Crit. Care Med. 37, 2827–2839 (2009).
Pucino, V., Bombardieri, M., Pitzalis, C. & Mauro, C. Lactate at the crossroads of metabolism, inflammation, and autoimmunity. Eur. J. Immunol. 47, 14–21 (2017).
Bergman, B. C. et al. Active muscle and whole body lactate kinetics after endurance training in men. J. Appl. Physiol. 87, 1684–1696 (1999).
Jin, N. et al. Identification of metabolic vulnerabilities of receptor tyrosine kinases-driven cancer. Nat. Commun. 10, 2701 (2019).
Brooks, G. A. Cell–cell and intracellular lactate shuttles. J. Physiol. 587, 5591–5600 (2009).
Tang, F. et al. Lactate-mediated glia–neuronal signalling in the mammalian brain. Nat. Commun. 5, 3284 (2014).
Colegio, O. R. et al. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature 513, 559–563 (2014).
Zhou, L. et al. Hepatitis B virus rigs the cellular metabolome to avoid innate immune recognition. Nat. Commun. 12, 98 (2021).
Doherty, J. R. et al. Blocking lactate export by inhibiting the Myc target MCT1 disables glycolysis and glutathione synthesis. Cancer Res. 74, 908–920 (2014).
Ždralević, M. et al. Disrupting the ‘Warburg effect’ re-routes cancer cells to OXPHOS offering a vulnerability point via ‘ferroptosis’-induced cell death. Adv. Biol. Regul. 68, 55–63 (2018).
Zong, Z. et al. Alanyl-tRNA synthetase, AARS1, is a lactate sensor and lactyltransferase that lactylates p53 and contributes to tumorigenesis. Cell 187, 2375–2392.e33 (2024).
Ju, J. et al. The alanyl-tRNA synthetase AARS1 moonlights as a lactyltransferase to promote YAP signaling in gastric cancer.J. Clin. Invest. 134, e174587 (2024).
Mao, Y. et al. Hypoxia induces mitochondrial protein lactylation to limit oxidative phosphorylation. Cell Res. 34, 13–30 (2024).
Li, H. et al. AARS1 and AARS2 sense l-lactate to regulate cGAS as global lysine lactyltransferases. Nature 634, 1229–1237 (2024).
Martinez-Outschoorn, U. E., Peiris-Pages, M., Pestell, R. G., Sotgia, F. & Lisanti, M. P. Cancer metabolism: a therapeutic perspective. Nat. Rev. Clin. Oncol. 14, 11–31 (2017).
Sharma, N. K. & Pal, J. K. Metabolic ink lactate modulates epigenomic landscape: a concerted role of pro-tumor microenvironment and macroenvironment during carcinogenesis. Curr. Mol. Med. 21, 177–181 (2021).
Warburg, O., Wind, F. & Negelein, E. The metabolism of tumors in the body. J. Gen. Physiol. 8, 519–530 (1927).
Vander Heiden, M. G., Cantley, L. C. & Thompson, C. B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 (2009).
Markert, C. L., Shaklee, J. B. & Whitt, G. S. Evolution of a gene. Multiple genes for LDH isozymes provide a model of the evolution of gene structure, function and regulation. Science 189, 102–114 (1975).
Claps, G. et al. The multiple roles of LDH in cancer. Nat. Rev. Clin. Oncol. 19, 749–762 (2022).
Jha, M. K., Lee, I. K. & Suk, K.Metabolic reprogramming by the pyruvate dehydrogenase kinase–lactic acid axis: linking metabolism and diverse neuropathophysiologies. Neurosci. Biobehav. Rev. 68, 1–19 (2016).
Li, X. et al. Lactate metabolism in human health and disease. Signal Transduct. Target. Ther. 7, 305 (2022).
Cai, T. Q. et al. Role of GPR81 in lactate-mediated reduction of adipose lipolysis. Biochem. Biophys. Res. Commun. 377, 987–991 (2008).
Liu, C. et al. Lactate inhibits lipolysis in fat cells through activation of an orphan G-protein-coupled receptor, GPR81. J. Biol. Chem. 284, 2811–2822 (2009).
Ahmed, K. et al. An autocrine lactate loop mediates insulin-dependent inhibition of lipolysis through GPR81. Cell Metab. 11, 311–319 (2010).
Liu, X. et al. Activation of GPR81 by lactate drives tumour-induced cachexia. Nat. Metab. 6, 708–723 (2024).
Feng, J. et al. Tumor cell-derived lactate induces TAZ-dependent upregulation of PD-L1 through GPR81 in human lung cancer cells. Oncogene 36, 5829–5839 (2017).
Yao, Z. et al. Dietary lactate supplementation protects against obesity by promoting adipose browning in mice. J. Agric. Food Chem. 68, 14841–14849 (2020).
Brown, T. P. et al. The lactate receptor GPR81 promotes breast cancer growth via a paracrine mechanism involving antigen-presenting cells in the tumor microenvironment. Oncogene 39, 3292–3304 (2020).
Chen, P. et al. Gpr132 sensing of lactate mediates tumor–macrophage interplay to promote breast cancer metastasis. Proc. Natl Acad. Sci. USA 114, 580–585 (2017).
Sun, J., Feng, Q., He, Y., Wang, M. & Wu, Y.Lactate activates CCL18 expression via H3K18 lactylation in macrophages to promote tumorigenesis of ovarian cancer. Acta Biochim. Biophys. Sin. 56, 1373–1386 (2024).
Luo, F. et al. Enhanced glycolysis, regulated by HIF-1α via MCT-4, promotes inflammation in arsenite-induced carcinogenesis. Carcinogenesis 38, 615–626 (2017).
Duan, Q. et al. Proton-coupled monocarboxylate transporters in cancer: from metabolic crosstalk, immunosuppression and anti-apoptosis to clinical applications. Front. Cell Dev. Biol. 10, 1069555 (2022).
Semenza, G. L. HIF-1: upstream and downstream of cancer metabolism. Curr. Opin. Genet. Dev. 20, 51–56 (2010).
Pucino, V., Cucchi, D. & Mauro, C.Lactate transporters as therapeutic targets in cancer and inflammatory diseases. Expert Opin. Ther. Targets 22, 735–743 (2018).
Li, H. et al. SLC5A8, a sodium transporter, is a tumor suppressor gene silenced by methylation in human colon aberrant crypt foci and cancers. Proc. Natl Acad. Sci. USA 100, 8412–8417 (2003).
Gopal, E. et al. Cloning and functional characterization of human SMCT2 (SLC5A12) and expression pattern of the transporter in kidney. Biochim. Biophys. Acta 1768, 2690–2697 (2007).
Haas, R. et al. Lactate regulates metabolic and pro-inflammatory circuits in control of T cell migration and effector functions. PLoS Biol. 13, e1002202 (2015).
Pucino, V. et al. Lactate buildup at the site of chronic inflammation promotes disease by inducing CD4+ T cell metabolic rewiring. Cell Metab. 30, 1055–1074.e8 (2019).
Romero, M. et al. Immunometabolic effects of lactate on humoral immunity in healthy individuals of different ages. Nat. Commun. 15, 7515 (2024).
Zhang, D. et al. Metabolic regulation of gene expression by histone lactylation. Nature 574, 575–580 (2019).
Gaffney, D. O. et al. Non-enzymatic lysine lactoylation of glycolytic enzymes. Cell Chem. Biol. 27, 206–213.e6 (2020).
Rabbani, N., Xue, M. & Thornalley, P. J. Activity, regulation, copy number and function in the glyoxalase system. Biochem. Soc. Trans. 42, 419–424 (2014).
Zhang, D. et al. Lysine l-lactylation is the dominant lactylation isomer induced by glycolysis. Nat. Chem. Biol. 21, 91–99 (2024).
Yu, J. et al. Histone lactylation drives oncogenesis by facilitating m6A reader protein YTHDF2 expression in ocular melanoma. Genome Biol. 22, 85 (2021).
Li, W. et al. Tumor-derived lactate promotes resistance to bevacizumab treatment by facilitating autophagy enhancer protein RUBCNL expression through histone H3 lysine 18 lactylation (H3K18la) in colorectal cancer. Autophagy 20, 114–130 (2024).
Xie, B. et al. CircXRN2 suppresses tumor progression driven by histone lactylation through activating the Hippo pathway in human bladder cancer. Mol. Cancer 22, 151 (2023).
Rho, H., Terry, A. R., Chronis, C. & Hay, N. Hexokinase 2-mediated gene expression via histone lactylation is required for hepatic stellate cell activation and liver fibrosis. Cell Metab. 35, 1406–1423.e8 (2023).
Xiong, J. et al. Lactylation-driven METTL3-mediated RNA m6A modification promotes immunosuppression of tumor-infiltrating myeloid cells. Mol. Cell 82, 1660–1677.e10 (2022).
Raychaudhuri, D. et al. Histone lactylation drives CD8+ T cell metabolism and function. Nat. Immunol. 25, 2140–2151 (2024).
Wang, N. et al. Histone lactylation boosts reparative gene activation post-myocardial infarction. Circ. Res. 131, 893–908 (2022).
Lin, X. et al. Augmentation of scleral glycolysis promotes myopia through histone lactylation. Cell Metab. 36, 511–525.e7 (2024).
Yang, L. et al. Nucleolin lactylation contributes to intrahepatic cholangiocarcinoma pathogenesis via RNA splicing regulation of MADD. J. Hepatol. 81, 651–666 (2024).
Pan, R. Y. et al. Positive feedback regulation of microglial glucose metabolism by histone H4 lysine 12 lactylation in Alzheimer’s disease. Cell Metab. 34, 634–648.e6 (2022).
Chen, H. et al. NBS1 lactylation is required for efficient DNA repair and chemotherapy resistance. Nature 631, 663–669 (2024).
Sun, L. et al. Lactylation of METTL16 promotes cuproptosis via m6A-modification on FDX1 mRNA in gastric cancer. Nat. Commun. 14, 6523 (2023).
Gu, J. et al. Tumor metabolite lactate promotes tumorigenesis by modulating MOESIN lactylation and enhancing TGF-β signaling in regulatory T cells. Cell Rep. 39, 110986 (2022).
Jin, J. et al. SIRT3-dependent delactylation of cyclin E2 prevents hepatocellular carcinoma growth. EMBO Rep. 24, e56052 (2023).
Broder, G. & Weil, M. H. Excess lactate: an index of reversibility of shock in human patients. Science 143, 1457–1459 (1964).
Yang, K. et al. Lactate promotes macrophage HMGB1 lactylation, acetylation, and exosomal release in polymicrobial sepsis. Cell Death Differ. 29, 133–146 (2022).
An, S. et al. PDHA1 hyperacetylation-mediated lactate overproduction promotes sepsis-induced acute kidney injury via Fis1 lactylation. Cell Death Dis. 14, 457 (2023).
Wang, X. et al. YY1 lactylation in microglia promotes angiogenesis through transcription activation-mediated upregulation of FGF2. Genome Biol. 24, 87 (2023).
Zhou, J. et al. Astrocytic LRP1 enables mitochondria transfer to neurons and mitigates brain ischemic stroke by suppressing ARF1 lactylation. Cell Metab. 36, 2054–2068.e14 (2024).
Yan, Q. et al. Lactylation of NAT10 promotes N4-acetylcytidine modification on tRNASer-CGA-1-1 to boost oncogenic DNA virus KSHV reactivation. Cell Death Differ. 31, 1362–1374 (2024).
Gao, R. et al. Mitochondrial pyruvate carrier 1 regulates fatty acid synthase lactylation and mediates treatment of nonalcoholic fatty liver disease. Hepatology 78, 1800–1815 (2023).
Zhang, X. et al. Screening, expression, purification and characterization of CoA-transferases for lactoyl-CoA generation. J. Ind. Microbiol. Biotechnol. 46, 899–909 (2019).
Dong, H. et al. YiaC and CobB regulate lysine lactylation in Escherichia coli. Nat. Commun. 13, 6628 (2022).
Liu, R. et al. Nuclear GTPSCS functions as a lactyl-CoA synthetase to promote histone lactylation and gliomagenesis. Cell Metab. 37, 377–394.e9 (2025).
Zhu, R. et al. ACSS2 acts as a lactyl-CoA synthetase and couples KAT2A to function as a lactyltransferase for histone lactylation and tumor immune evasion. Cell Metab. 37, 361–376.e7 (2025).
Trujillo, M. N. et al. Lactoylglutathione promotes inflammatory signaling in macrophages through histone lactoylation. Mol. Metab. 81, 101888 (2024).
Zhao, S., Zhang, X. & Li, H. Beyond histone acetylation—writing and erasing histone acylations. Curr. Opin. Struct. Biol. 53, 169–177 (2018).
Sun, L., Zhang, H. & Gao, P. Metabolic reprogramming and epigenetic modifications on the path to cancer. Protein Cell 13, 877–919 (2022).
Cui, H. et al. Lung myofibroblasts promote macrophage profibrotic activity through lactate-induced histone lactylation. Am. J. Respir. Cell Mol. Biol. 64, 115–125 (2021).
Niu, Z. et al. HBO1 catalyzes lysine lactylation and mediates histone H3K9la to regulate gene transcription. Nat. Commun. 15, 3561 (2024).
Xie, B. et al. KAT8-catalyzed lactylation promotes eEF1A2-mediated protein synthesis and colorectal carcinogenesis. Proc. Natl Acad. Sci. USA 121, e2314128121 (2024).
Akella, J. S. et al. MEC-17 is an α-tubulin acetyltransferase. Nature 467, 218–222 (2010).
Shida, T., Cueva, J. G., Xu, Z., Goodman, M. B. & Nachury, M. V. The major α-tubulin K40 acetyltransferase αTAT1 promotes rapid ciliogenesis and efficient mechanosensation. Proc. Natl Acad. Sci. USA 107, 21517–21522 (2010).
Sun, S. et al. Metabolic regulation of cytoskeleton functions by HDAC6-catalyzed α-tubulin lactylation. Nat. Commun. 15, 8377 (2024).
Christensen, D. G. et al. Identification of novel protein lysine acetyltransferases in Escherichia coli. mBio 9, e01905–e01918 (2018).
Liu, X. et al. The structural basis of protein acetylation by the p300/CBP transcriptional coactivator. Nature 451, 846–850 (2008).
Kaczmarska, Z. et al. Structure of p300 in complex with acyl-CoA variants. Nat. Chem. Biol. 13, 21–29 (2017).
Wang, Y. et al. KAT2A coupled with the α-KGDH complex acts as a histone H3 succinyltransferase. Nature 552, 273–277 (2017).
Ringel, A. E. & Wolberger, C. Structural basis for acyl-group discrimination by human Gcn5L2. Acta Crystallogr. D Struct. Biol. 72, 841–848 (2016).
Han, Z. et al. Revealing the protein propionylation activity of the histone acetyltransferase MOF (males absent on the first). J. Biol. Chem. 293, 3410–3420 (2018).
Varner, E. L. et al. Quantification of lactoyl-CoA (lactyl-CoA) by liquid chromatography mass spectrometry in mammalian cells and tissues. Open Biol. 10, 200187 (2020).
Wishart, D. S. et al. HMDB: the Human Metabolome Database. Nucleic Acids Res. 35, D521–D526 (2007).
Matsumoto, K. et al. In vitro analysis of d-lactyl-CoA-polymerizing polyhydroxyalkanoate synthase in polylactate and poly(lactate-co-3-hydroxybutyrate) syntheses. Biomacromolecules 19, 2889–2895 (2018).
Sabari, B. R., Zhang, D., Allis, C. D. & Zhao, Y.Metabolic regulation of gene expression through histone acylations. Nat. Rev. Mol. Cell Biol. 18, 90–101 (2017).
Shvedunova, M. & Akhtar, A. Modulation of cellular processes by histone and non-histone protein acetylation. Nat. Rev. Mol. Cell Biol. 23, 329–349 (2022).
Sun, L. et al. Evolutionary gain of alanine mischarging to noncognate tRNAs with a G4:U69 base pair. J. Am. Chem. Soc. 138, 12948–12955 (2016).
Sun, L., Song, Y., Blocquel, D., Yang, X. L. & Schimmel, P. Two crystal structures reveal design for repurposing the C-Ala domain of human AlaRS. Proc. Natl Acad. Sci. USA 113, 14300–14305 (2016).
Han, J. M. et al. Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell 149, 410–424 (2012).
He, X. D. et al. Sensing and transmitting intracellular amino acid signals through reversible lysine aminoacylations. Cell Metab. 27, 151–166.e6 (2018).
D’Hulst, G. et al. PHD1 controls muscle mTORC1 in a hydroxylation-independent manner by stabilizing leucyl tRNA synthetase. Nat. Commun. 11, 174 (2020).
Zhou, Q. et al. Phenylalanine impairs insulin signaling and inhibits glucose uptake through modification of IRβ. Nat. Commun. 13, 4291 (2022).
Brizel, D. M. et al. Elevated tumor lactate concentrations predict for an increased risk of metastases in head-and-neck cancer. Int. J. Radiat. Oncol. Biol. Phys. 51, 349–353 (2001).
Carter, C. W. JrCognition, mechanism, and evolutionary relationships in aminoacyl-tRNA synthetases. Annu. Rev. Biochem. 62, 715–748 (1993).
Beebe, K., Ribas De Pouplana, L. & Schimmel, P. Elucidation of tRNA-dependent editing by a class II tRNA synthetase and significance for cell viability. EMBO J. 22, 668–675 (2003).
Lee, J. W. et al. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature 443, 50–55 (2006).
Guo, M. et al. Paradox of mistranslation of serine for alanine caused by AlaRS recognition dilemma. Nature 462, 808–812 (2009).
Kamagata, K. et al. Liquid-like droplet formation by tumor suppressor p53 induced by multivalent electrostatic interactions between two disordered domains. Sci. Rep. 10, 580 (2020).
Chen, C., Fu, G., Guo, Q., Xue, S. & Luo, S. Z. Phase separation of p53 induced by its unstructured basic region and prevented by oncogenic mutations in tetramerization domain. Int. J. Biol. Macromol. 222, 207–216 (2022).
Chen, Q., Wu, Y., Dai, Z., Zhang, Z. & Yang, X. Phosphorylation and specific DNA improved the incorporation ability of p53 into functional condensates. Int. J. Biol. Macromol. 230, 123221 (2023).
Dai, Z., Li, G., Chen, Q. & Yang, X. Ser392 phosphorylation modulated a switch between p53 and transcriptional condensates. Biochim. Biophys. Acta Gene Regul. Mech. 1865, 194827 (2022).
Yang, Z. et al. Lactylome analysis suggests lactylation-dependent mechanisms of metabolic adaptation in hepatocellular carcinoma. Nat. Metab. 5, 61–79 (2023).
Kalderon, D., Roberts, B. L., Richardson, W. D. & Smith, A. E. A short amino acid sequence able to specify nuclear location. Cell 39, 499–509 (1984).
Wang, J., Wang, Z., Wang, Q., Li, X. & Guo, Y. Ubiquitous protein lactylation in health and diseases. Cell. Mol. Biol. Lett. 29, 23 (2024).
Chen, Y. et al. Metabolic regulation of homologous recombination repair by MRE11 lactylation. Cell 187, 294–311 e221 (2024).
Sun, T. et al. Oxamate enhances the efficacy of CAR-T therapy against glioblastoma via suppressing ectonucleotidases and CCR8 lactylation. J. Exp. Clin. Cancer Res. 42, 253 (2023).
Xu, K., Zhang, K., Wang, Y. & Gu, Y.Comprehensive review of histone lactylation: structure, function, and therapeutic targets. Biochem. Pharmacol. 225, 116331 (2024).
Yu, X. et al. Histone lactylation: from tumor lactate metabolism to epigenetic regulation. Int. J. Biol. Sci. 20, 1833–1854 (2024).
Zhang, Y., Song, H., Li, M. & Lu, P. Histone lactylation bridges metabolic reprogramming and epigenetic rewiring in driving carcinogenesis: oncometabolite fuels oncogenic transcription. Clin. Transl. Med. 14, e1614 (2024).
Qu, J., Li, P. & Sun, Z.Histone lactylation regulates cancer progression by reshaping the tumor microenvironment. Front. Immunol. 14, 1284344 (2023).
Li, J. et al. Lactate regulates major zygotic genome activation by H3K18 lactylation in mammals. Natl Sci. Rev. 11, nwad295 (2024).
Dai, S. K. et al. Dynamic profiling and functional interpretation of histone lysine crotonylation and lactylation during neural development. Development 149, dev200049 (2022).
Merkuri, F., Rothstein, M. & Simoes-Costa, M. Histone lactylation couples cellular metabolism with developmental gene regulatory networks. Nat. Commun. 15, 90 (2024).
Li, L. et al. Glis1 facilitates induction of pluripotency via an epigenome–metabolome–epigenome signalling cascade. Nat. Metab. 2, 882–892 (2020).
Yue, Q. et al. Histone H3K9 lactylation confers temozolomide resistance in glioblastoma via LUC7L2-mediated MLH1 intron retention. Adv. Sci. 11, e2309290 (2024).
Wan, N. et al. Cyclic immonium ion of lactyllysine reveals widespread lactylation in the human proteome. Nat. Methods 19, 854–864 (2022).
Yang, D. et al. Identification of lysine-lactylated substrates in gastric cancer cells. iScience 25, 104630 (2022).
Duan, Y. et al. Integrated lactylome characterization reveals the molecular dynamics of protein regulation in gastrointestinal cancers. Adv. Sci. 11, e2400227 (2024).
Amorini, A. M. et al. Serum lactate as a novel potential biomarker in multiple sclerosis. Biochim. Biophys. Acta 1842, 1137–1143 (2014).
Hsu, Y. C. & Hsu, C. W. Septic acute kidney injury patients in emergency department: the risk factors and its correlation to serum lactate. Am. J. Emerg. Med. 37, 204–208 (2019).
Jia, M. et al. ULK1-mediated metabolic reprogramming regulates Vps34 lipid kinase activity by its lactylation. Sci. Adv. 9, eadg4993 (2023).
Wang, J. et al. Lactylation of PKM2 suppresses inflammatory metabolic adaptation in pro-inflammatory macrophages. Int. J. Biol. Sci. 18, 6210–6225 (2022).
De Ruijter, A. J., van Gennip, A. H., Caron, H. N., Kemp, S. & van Kuilenburg, A. B. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem. J. 370, 737–749 (2003).
Zu, H. et al. SIRT2 functions as a histone delactylase and inhibits the proliferation and migration of neuroblastoma cells. Cell Discov. 8, 54 (2022).
Jennings, E. Q. et al. Sirtuin 2 regulates protein lactoylLys modifications. ChemBioChem 22, 2102–2106 (2021).
Moreno-Yruela, C. et al. Class I histone deacetylases (HDAC1-3) are histone lysine delactylases. Sci. Adv. 8, eabi6696 (2022).
Millard, C. J., Watson, P. J., Fairall, L. & Schwabe, J. W. R. Targeting class I histone deacetylases in a “complex” environment. Trends Pharmacol. Sci. 38, 363–377 (2017).
Zessin, M. et al. Uncovering robust delactoylase and depyruvoylase activities of HDAC isoforms. ACS Chem. Biol. 17, 1364–1375 (2022).
Du, R. et al. Sirtuin 1/sirtuin 3 are robust lysine delactylases and sirtuin 1-mediated delactylation regulates glycolysis. iScience 27, 110911 (2024).
Zhang, N. et al. α-myosin heavy chain lactylation maintains sarcomeric structure and function and alleviates the development of heart failure. Cell Res. 33, 679–698 (2023).
Bell, E. L. & Guarente, L. The SirT3 divining rod points to oxidative stress. Mol. Cell 42, 561–568 (2011).
Dai, W. et al. Lactate promotes myogenesis via activating H3K9 lactylation-dependent up-regulation of Neu2 expression. J. Cachexia Sarcopenia Muscle 14, 2851–2865 (2023).
Guo, Z. et al. Natural product fargesin interferes with H3 histone lactylation via targeting PKM2 to inhibit non-small cell lung cancer tumorigenesis. Biofactors 50, 592–607 (2024).
Wang, Y. et al. Novel strategies to improve tumour therapy by targeting the proteins MCT1, MCT4 and LAT1. Eur. J. Med. Chem. 226, 113806 (2021).
Tasdogan, A. et al. Metabolic heterogeneity confers differences in melanoma metastatic potential. Nature 577, 115–120 (2020).
Sisignano, M., Fischer, M. J. M. & Geisslinger, G. Proton-sensing GPCRs in health and disease. Cells 10, 2050 (2021).
Obinata, H. & Izumi, T. G2A as a receptor for oxidized free fatty acids. Prostaglandins Other Lipid Mediat. 89, 66–72 (2009).
Damaghi, M., Wojtkowiak, J. W. & Gillies, R. J. pH sensing and regulation in cancer. Front. Physiol. 4, 370 (2013).
Lasko, L. M. et al. Discovery of a selective catalytic p300/CBP inhibitor that targets lineage-specific tumours. Nature 550, 128–132 (2017).
Saunders, B. et al. β-alanine supplementation to improve exercise capacity and performance: a systematic review and meta-analysis. Br. J. Sports Med. 51, 658–669 (2017).
Acknowledgements
The current work was supported by the Chinese National Natural Science Fund (31925013, 32125016, T2321005, U20A20393, 32070907, 31871405, 31870902, W2411011 and U24A20371), a special program of the Ministry of Science and Technology of China (2024YFC2707400, 2021YFA1101000, 2022YFA1105200 and 2023YFA1800200), a Key Research and Development Program of Zhejiang Province (2024C03142), a joint project of the Pinnacle Disciplinary Group, the Second Affiliated Hospital of Chongqing Medical University, the Science Foundation of Jiangsu Province (19KJA550003), the Suzhou Innovation and Entrepreneurship Leading Talent Program (ZXL2022505), a Suzhou Medical College Basic Frontier Innovation cross-research project (YXY2303027), key cross-research projects of the School of Medicine at Soochow University (YXY2303027) and the Suzhou International Joint Laboratory for Diagnosis and Treatment of Brain Diseases. We apologize to those researchers whose related work we were not able to cite in this Perspective. Figures were created using BioRender.com.
Author information
Authors and Affiliations
Contributions
Z.Z. conceived of and drafted the manuscript and produced the figures. J.R., B.Y., L.Z. and F.Z. discussed the concepts of the manuscript, provided valuable discussion and revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Cell Biology thanks Claudio Mauro, James Galligan and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Tables 1 and 2
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zong, Z., Ren, J., Yang, B. et al. Emerging roles of lysine lactyltransferases and lactylation. Nat Cell Biol 27, 563–574 (2025). https://doi.org/10.1038/s41556-025-01635-8
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41556-025-01635-8
This article is cited by
-
Glucose metabolism and its direct action in cancer and immune regulation: opportunities and challenges for metabolic targeting
Journal of Biomedical Science (2025)
-
Histone and non-histone lactylation: molecular mechanisms, biological functions, diseases, and therapeutic targets
Molecular Biomedicine (2025)
-
Irinotecan alleviates chemoresistance to anthracyclines through the inhibition of AARS1-mediated BLM lactylation and homologous recombination repair
Signal Transduction and Targeted Therapy (2025)
-
A systematic review of protein post-translational modifications in sepsis
Molecular Biology Reports (2025)
-
Tackling tumor hypoxia: advances in breaking the oncogenic HIF-1α–p300/CBP alliance
Investigational New Drugs (2025)