Fig. 1: Derivation and characterization of ellNSCs and allNSCs.
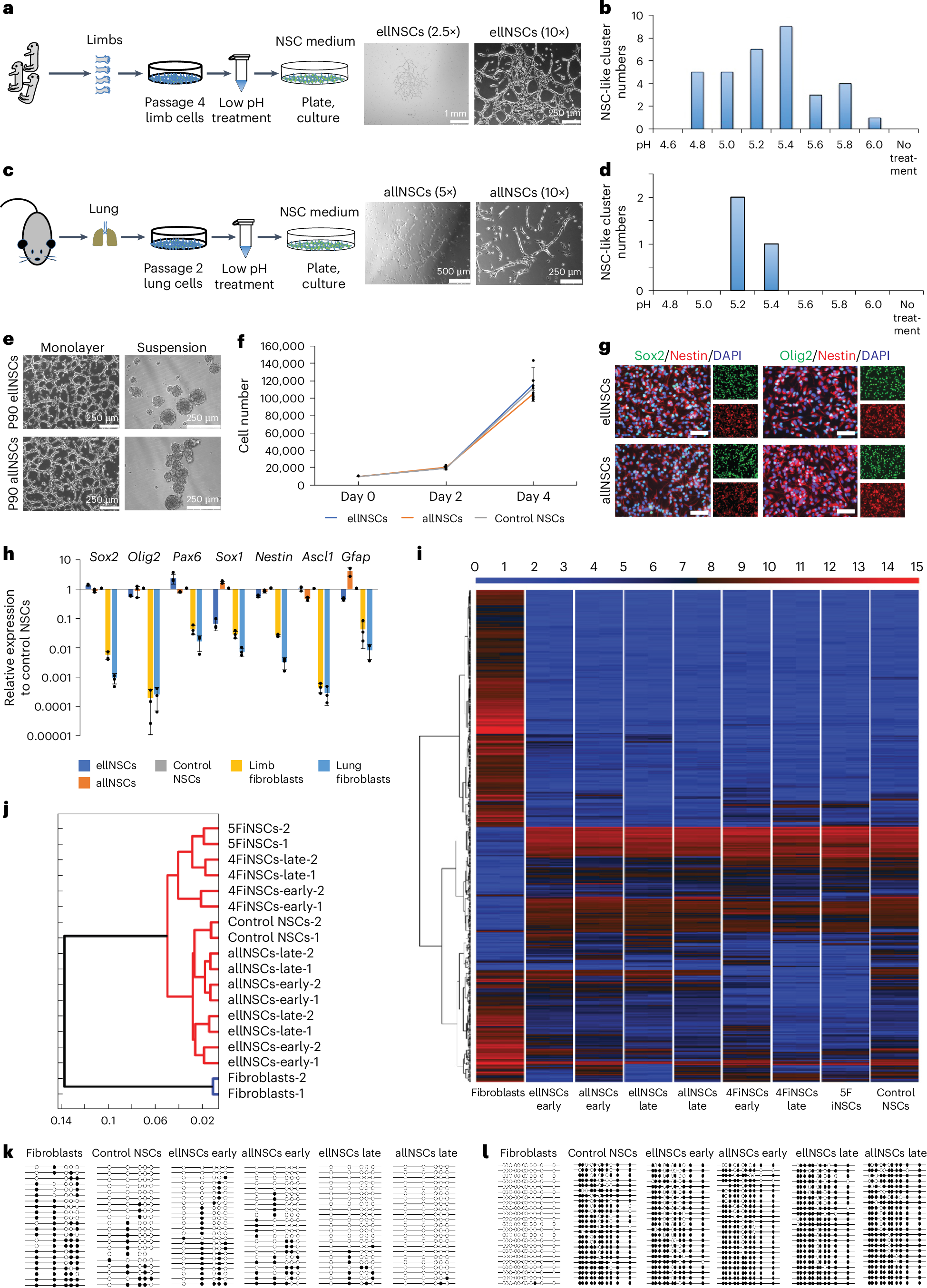
a, Schematic for deriving ellNSCs and morphology of early cluster. b, Number of NSC-like clusters obtained with different pH conditions from passage-4 embryonic limb cells; n = 12. c, Schematic for deriving allNSCs and morphology of early cluster. d, Number of NSC-like clusters obtained with different pH conditions from passage-2 adult lung cells; n = 10. e, LdNSCs stably maintained for 90 passages, representative of three biological replicates. f, Proliferation rates of passage-90 ldNSCs and control NSCs. The data represent mean ± s.d. (n = 3 biological replicates). g, Immunofluorescence of NSC markers in ellNSCs and allNSCs, representative of three biological replicates. Scale bar, 50 μm. h, RT–qPCR of NSC markers in samples. All data are calibrated to brain NSCs, whose expression is considered to be 1 for all genes. The data represent mean ± s.d. (n = 3 biological replicates). i, Heatmap of global gene expression across samples. Colour bar indicates gene expression in log2 scale. n = 2 biological replicates, all replicates are shown. j, Hierarchical clustering of the cell lines based on i. k,l, Bisulfite sequencing PCR on the second intron of Nestin (k) and promoter of Col1a1 (l) in samples. Open and filled circles represent unmethylated and methylated CpGs, respectively.
