Abstract
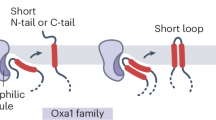
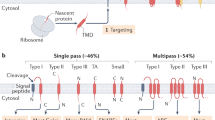
Mitochondria are critical double-membraned organelles that act as biosynthetic and bioenergetic cellular factories, with the outer membrane providing an interface with the rest of the cell. Mitochondrial outer membrane proteins regulate a variety of processes, including metabolism, innate immunity and apoptosis. Although the biophysical and functional diversity of these proteins is highly documented, the mechanisms of their biogenesis and the integration of that into cellular homeostasis are just starting to take shape. Here, focusing on α-helical outer membrane proteins, we review recent insights into the mechanisms of synthesis and cytosolic chaperoning, insertion and assembly in the lipid bilayer, and quality control of unassembled or mislocalized transmembrane domains. We further discuss the role convergent evolution played in this process, comparing key biogenesis players from lower eukaryotes, including yeast and trypanosomes, with multicellular metazoan systems, and draw comparisons with the endoplasmic reticulum biogenesis system, in which membrane proteins face similar challenges.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Von Heijne, G. The membrane protein universe: what’s out there and why bother? J. Intern. Med. 261, 543–557 (2007).
Hegde, R. S. & Keenan, R. J. The mechanisms of integral membrane protein biogenesis. Nat. Rev. Mol. Cell Biol. 23, 107–124 (2022).
Rapoport, T. A., Li, L. & Park, E. Structural and mechanistic insights into protein translocation. Annu. Rev. Cell Dev. Biol. 33, 369–390 (2017).
Wickner, W. & Schekman, R. Protein translocation across biological membranes. Science 310, 1452–1456 (2005).
Rapoport, T. A. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature 450, 663–669 (2007).
White, S. H. & von Heijne, G. How translocons select transmembrane helices. Annu. Rev. Biophys. 37, 23–42 (2008).
Friedman, J. R. & Nunnari, J. Mitochondrial form and function. Nature 505, 335–343 (2014).
Guna, A. & Hegde, R. S. Transmembrane domain recognition during membrane protein biogenesis and quality control. Curr. Biol. 28, R498–R511 (2018).
Becker, T., Song, J. & Pfanner, N. Versatility of preprotein transfer from the cytosol to mitochondria. Trends Cell. Biol. 7, 534–548 (2019).
Gupta, A. & Becker, T. Mechanisms and pathways of mitochondrial outer membrane protein biogenesis. Biochem. Biophys. Acta Bioenerg. 1862, 148323 (2021).
Wiedemann, N. et al. Machinery for protein sorting and assembly in the mitochondrial outer membrane. Nature 424, 565–571 (2003).
Paschen, S. A. et al. Evolutionary conservation of biogenesis of β-barrel membrane proteins. Nature 426, 862–866 (2003).
Höhr, A. I. C. et al. Membrane protein insertion through a mitochondrial β-barrel gate. Science 359, eaah6834 (2018).
Diederichs, K. A. et al. Structural insight into mitochondrial β-barrel outer membrane protein biogenesis. Nat. Commun. 11, 3290 (2020).
Muthukumar, G. et al. Triaging of α-helical proteins to the mitochondrial outer membrane by distinct chaperone machinery based on substrate topology. Mol. Cell 84, 1101–1119.e9 (2024).
Drwesh, L. et al. A network of cytosolic (co)chaperones promotes the biogenesis of mitochondrial signal-anchored outer membrane proteins. eLife 11, e77706 (2022).
Doan, K. N. et al. The mitochondrial import complex MIM functions as main translocase for a-helical outer membrane proteins. Cell Rep. 31, 107567 (2020).
Vitali, D. G. et al. Independent evolution of functionally exchangeable mitochondrial outer membrane import complexes. eLife 7, e34488 (2018).
Guna, A. et al. MTCH2 is a mitochondrial outer membrane protein insertase. Science 378, 317–322 (2022).
Bykov, Y. S., Rapaport, D., Herrmann, J. M. & Schuldiner, M. Cytosolic events in the biogenesis of mitochondrial proteins. Trends Biochem. Sci. 45, 650–667 (2020).
Kellems, R. E., Allison, V. F. & Butow, R. A. Cytoplasmic type 80 S ribosomes associated with yeast mitochondria. II. Evidence for the association of cytoplasmic ribosomes with the outer mitochondrial membrane in situ. J. Biol. Chem. 249, 3297–3303 (1974).
Kellems, R. E., Allison, V. F. & Butow, R. A. Cytoplasmic type 80S ribosomes associated with yeast mitochondria. IV. Attachment of ribosomes to the outer membrane of isolated mitochondria. J. Cell Biol. 65, 1–14 (1975).
Gold, V. A., Chroscicki, P., Bragoszewski, P. & Chacinska, A. Visualization of cytosolic ribosomes on the surface of mitochondria by electron cryo-tomography. EMBO Rep. 18, 1786–1800 (2017).
Fazal, F. M. et al. Atlas of subcellular RNA localization revealed by APEX-seq. Cell 178, 473–490 (2019).
Williams, C. C., Jan, C. H. & Weissman, J. S. Targeting and plasticity of mitochondrial proteins revealed by proximity-specific ribosome profiling. Science 346, 748–751 (2014).
Deshaies, R. J., Koch, B. D., Werner-Washburne, M., Craig, E. A. & Schekman, R. A subfamily of stress proteins facilitates translocation of secretory and mitochondrial precursor polypeptides. Nature 332, 800–805 (1988).
Young, J. C., Hoogenraad, N. J. & Hartl, F. U. Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell 112, 41–50 (2003).
Fan, A. C. Y., Bhangoo, M. K. & Young, J. C. Hsp90 functions in the targeting and outer membrane translocation steps of Tom70-mediated mitochondrial import. J. Biol. Chem. 28, 33313–33324 (2006).
Bloss, T. A., Witze, E. S. & Rothman, J. H. Suppression of CED-3-independent apoptosis by mitochondrial βNAC in Caenorhabditis elegans. Nature 424, 1066–1071 (2003).
Markesich, D. C., Gajewski, K. M., Nazimiec, M. E. & Beckingham, K. bicaudal encodes the Drosophila beta NAC homolog, a component of the ribosomal translational machinery. Development 127, 559–572 (2000).
Fünfschilling, U. & Rospert, S. Nascent polypeptide-associated complex stimulates protein import into yeast mitochondria. Mol. Biol. Cell. 10, 3289–3299 (1999).
Lesnik, C., Cohen, Y., Atir-Lande, A., Schuldiner, M. & Arava, Y. OM14 is a mitochondrial receptor for cytosolic ribosomes that supports co-translational import into mitochondria. Nat. Commun. 5, 5711 (2014).
Ponce-Rojas, J. C. et al. αβ’-NAC cooperates with Sam37 to mediate early stages of mitochondrial protein import. FEBS J. 284, 814–830 (2017).
Lauring, B., Sakai, H., Kreibich, G. & Wiedmann, M. Nascent polypeptide-associated complex protein prevents mistargeting of nascent chains to the endoplasmic reticulum. Proc. Natl Acad. Sci. USA 92, 5411–5415 (1995).
Gamerdinger, M., Hanebuth, M. A., Frickey, T. & Deuerling, E. The principle of antagonism ensures protein targeting specificity at the endoplasmic reticulum. Science 348, 201–207 (2015).
Jomaa, A. et al. Mechanism of signal sequence handover from NAC to SRP on ribosomes during ER-protein targeting. Science 375, 839–844 (2022).
Sheffield, W. P., Shore, G. C. & Randall, S. K. Mitochondrial precursor protein. Effects of 70-kilodalton heat shock protein on polypeptide folding, aggregation, and import competence. J. Biol. Chem. 265, 11069–11076 (1990).
Jores, T. et al. Cytosolic Hsp70 and Hsp40 chaperones enable the biogenesis of mitochondrial β-barrel proteins. J. Cell Biol. 217, 30913108 (2018).
Hoseini, H. et al. The cytosolic cochaperone Sti1 is relevant for mitochondrial biogenesis and morphology. FEBS J. 283, 3338–3352 (2016).
Zara, V., Ferramosca, A., Robitaille-Foucher, P., Palmieri, F. & Young, J. C. Mitochondrial carrier protein biogenesis: role of the chaperones Hsc70 and Hsp90. Biochem. J. 419, 369–375 (2009).
Bhangoo, M. K. et al. Multiple 40-kDa heat-shock protein chaperones function in Tom70-dependent mitochondrial import. Mol. Biol. Cell. 18, 3414–3428 (2007).
Artigues, A., Iriarte, A. & Martinez-Carrion, M. Binding to chaperones allows import of a purified mitochondrial precursor into mitochondria. J. Biol. Chem. 277, 25047–25055 (2002).
Caplan, A. J., Cyr, D. M. & Douglas, M. G. YDJ1p facilitates polypeptide translocation across different intracellular membranes by a conserved mechanism. Cell 71, 1143–1155 (1992).
Kanazawa, M., Terada, K., Kato, S. & Mori, M. HSDJ, a human homolog of DnaJ, is farnesylated and is involved in protein import into mitochondria. J. Biochem. 121, 890–895 (1997).
Kampinga, H. H. & Craig, E. A. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 11, 579–592 (2010).
Przybyla, L. & Gilbert, L. A. A new era in functional genomics screens. Nat. Rev. Genet. 23, 89–103 (2022).
Bock, C. et al. High-content CRISPR screening. Nat. Rev. Methods Prim. 2, 8 (2022).
Lotz, G. P., Brychzy, A., Heinz, S. & Obermann, W. M. A novel HSP90 chaperone complex regulates intracellular vesicle transport. J. Cell Sci. 121, 717–723 (2008).
Philp, L. K. et al. SGTA: a new player in the molecular co-chaperone game. Horm. Cancer 4, 343–357 (2013).
Cho, H. & Shan, S. O. Substrate relay in an Hsp70-cochaperone cascade safeguards tail-anchored membrane protein targeting. EMBO J. 37, e99264 (2018).
Abe, Y. et al. Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20. Cell 100, 551–560 (2000).
Su, J. et al. Structural basis of Tom20 and Tom22 cytosolic domains as the human TOM complex receptors. Proc. Natl Acad. Sci. USA 119, e2200158119 (2022).
Hill, K. et al. Tom40 forms the hydrophilic channel of the mitochondrial import pore for preproteins. Nature 395, 516–521 (1998).
Tucker, K. & Park, E. Cryo-EM structure of the mitochondrial protein-import channel TOM complex at near-atomic resolution. Nat. Struct. Mol. Biol. 12, 1158–1166 (2019).
Wiedemann, N. & Pfanner, N. Mitochondrial machineries for protein import and assembly. Annu. Rev. Biochem. 86, 685–714 (2017).
Kutik, S. et al. Dissecting membrane insertion of mitochondrial β-barrel proteins. Cell 132, 1011–1024 (2008).
Walther, D. M. & Rapaport, D. Biogenesis of mitochondrial outer membrane proteins. Biochem. Biophys. Acta 1793, 42–51 (2009).
Becker, T. et al. The mitochondrial import protein Mim1 promotes biogenesis of multispanning outer membrane proteins. J. Cell Biol. 194, 387–395 (2011).
Krüger, V. et al. Identification of new channels by systematic analysis of the mitochondrial outer membrane. J. Cell Biol. 216, 3485–3495 (2017).
Käser, S. et al. Outer membrane protein functions as integrator of protein import and DNA inheritance in mitochondria. Proc. Natl Acad. Sci. USA 113, E4467–E4475 (2016).
Dimmer, K. S. et al. A crucial role for Mim2 in the biogenesis of mitochondrial outer membrane proteins. J. Cell Sci. 125, 3464–3473 (2012).
Lueder, F. & Lithgow, T. The three domains of the mitochondrial outer membrane protein Mim1 have discrete functions in assembly of the TOM complex. FEBS Lett. 583, 1475–1480 (2009).
Busch, J. D., Fielden, L. F., Pfanner, N. & Wiedemann, N. Mitochondrial protein transport: versatility of translocases and mechanisms. Mol. Cell 83, 890–910 (2023).
Ruprecht, J. J. & Kunji, E. R. S. The SLC25 mitochondrial carrier family: structure and mechanism. Trends Biochem. Sci. 45, 244–258 (2020).
Pleiner, T. et al. Structural basis for membrane insertion by the human ER membrane protein complex. Science 369, 433–436 (2020).
Wu, X. et al. Structural basis of ER-associated protein degradation mediated by the Hrd1 ubiquitin ligase complex. Science 368, eaaz2449 (2020).
Kumazaki, K. et al. Structural basis of Sec-independent membrane protein insertion by YidC. Nature 509, 516–520 (2014).
Dimogkioka, A. R., Elias, A. & Rapaport, D. The mammalian protein MTCH1 can function as an insertase. Preprint at bioRxiv https://doi.org/10.1101/2024.12.11.627916 (2024).
Labbé, K. et al. The modified mitochondrial outer membrane carrier MTCH2 links mitochondrial fusion to lipogenesis. J. Cell Biol. 220, e202103122 (2021).
Rottiers, V. et al. MTCH2 is a conserved regulator of lipid homeostasis. Obesity 25, 616–625 (2017).
Chitwood, P. J. & Hegde, R. S. An intramembrane chaperone complex facilitates membrane protein biogenesis. Nature 584, 630–634 (2020).
Brix, J., Dietmeier, K. & Pfanner, N. Differential recognition of preproteins by the purified cytosolic domains of the mitochondrial import receptors Tom20, Tom22, and Tom70. J. Biol. Chem. 272, 20730–20735 (1997).
Van Wilpe, S. et al. Tom22 is a multifunctional organizer of the mitochondrial preprotein translocase. Nature 401, 485–489 (1999).
Yamano, K. et al. Tom20 and Tom22 share the common signal recognition pathway in mitochondrial protein import. J. Biol. Chem. 283, 3799–3807 (2008).
Brix, J. et al. The mitochondrial import receptor Tom70: identification of a 25 kDa core domain with a specific binding site for preproteins. J. Mol. Biol. 303, 479–488 (2000).
Backes, S. et al. Tom70 enhances mitochondrial preprotein import efficiency by binding to internal targeting sequences. J. Cell Biol. 217, 1369–1382 (2018).
Backes, S. et al. The chaperone-binding activity of the mitochondrial surface receptor Tom70 protects the cytosol against mitoprotein-induced stress. Cell Rep. 35, 108936 (2021).
Kato, H., Lu, Q., Rapaport, D. & Kozjak-Pavlovic, V. Tom70 is essential for PINK1 import into mitochondria. PLoS ONE 8, e58435 (2013).
Vance, D. E., Choy, P. C., Farren, S. B., Lim, P. H. & Schneider, W. J. Asymmetry of phospholipid biosynthesis. Nature 270, 268–269 (1977).
Vance, J. E. Phospholipid synthesis and transport in mammalian cells. Traffic 16, 1–18 (2015).
Reinisch, K. M. & Prinz, W. A. Mechanisms of nonvesicular lipid transport. J. Cell Biol. 220, e202012058 (2021).
Pomorski, T. & Menon, A. K. Lipid flippases and their biological functions. Cell. Mol. Life Sci. 63, 2908–2921 (2006).
Bethel, N. P. & Grabe, M. Atomistic insight into lipid translocation by a TMEM16 scramblase. Proc. Natl Acad. Sci. USA 113, 14049–14054 (2016).
Li, D., Rocha-Roa, C., Schilling, M. A., Reinisch, K. M. & Vanni, S. Lipid scrambling is a general feature of protein insertases. Proc. Natl Acad. Sci. USA 121, e2319476121 (2024).
Itakura, E. et al. Ubiquilins chaperone and triage mitochondrial membrane proteins for degradation. Mol. Cell 63, 21–33 (2016).
Deng, H. X. et al. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature 477, 211–215 (2011).
Rodrigo-Brenni, M. C., Gutierrez, E. & Hegde, R. S. Cytosolic quality control of mislocalized proteins requires RNF126 recruitment to Bag6. Mol. Cell 55, 227–237 (2014).
Shao, S., Rodrigo-Brenni, M. C., Kivlen, M. H. & Hegde, R. S. Mechanistic basis for a molecular triage reaction. Science 355, 298–302 (2017).
Wohlever, M. L., Mateja, A., McGilvray, P. T., Day, K. J. & Keenan, R. J. Msp1 Is a membrane protein dislocase for tail-anchored proteins. Mol. Cell 67, 194–202.e6 (2017).
Matsumoto, S. et al. Msp1 clears mistargeted proteins by facilitating their transfer from mitochondria to the ER. Mol. Cell 76, 191–205.e10 (2019).
Wang, L., Toutkoushian, H., Belyy, V., Kokontis, C. Y. & Walter, P. Conserved structural elements specialize ATAD1 as a membrane protein extraction machine. eLife 11, e73941 (2022).
Kim, J. et al. ATAD1 prevents clogging of TOM and damage caused by un-imported mitochondrial proteins. Cell Rep. 43, 114473 (2024).
Wang, L. & Walter, P. Msp1/ATAD1 in protein quality control and regulation of synaptic activities. Annu. Rev. Cell Dev. Biol. 36, 141–164 (2020).
Weir, N. R., Kamber, R. A., Martenson, J. S. & Denic, V. The AAA protein Msp1 mediates clearance of excess tail-anchored proteins from the peroxisomal membrane. eLife 6, e28507 (2017).
Cichocki, B. A., Krumpe, K., Vitali, D. G. & Rapaport, D. Pex19 is involved in importing dually targeted tail-anchored proteins to both mitochondria and peroxisomes. Traffic 19, 770–785 (2018).
Aravindan, N. et al. Mpf1 is a novel factor that affects the dual distribution of tail-anchored proteins between mitochondria and peroxisomes. EMBO Rep. https://doi.org/10.1038/s44319-025-00440-6 (2025).
Nagashima, S., Tokuyama, T., Yonashiro, R., Inatome, R. & Yanagi, S. Roles of mitochondrial ubiquitin ligase MITOL/MARCH5 in mitochondrial dynamics and diseases. J. Biochem. 155, 273–279 (2014).
McKenna, M. J. et al. The endoplasmic reticulum P5A-ATPase is a transmembrane helix dislocase. Science 369, eabc5809 (2020).
McKenna, M. J., Adams, B. M., Chu, V., Paulo, J. A. & Shao, S. ATP13A1 prevents ERAD of folding-competent mislocalized and misoriented proteins. Mol. Cell 82, 4277–4289 (2022).
Mun, S. H. et al. Marchf6 E3 ubiquitin ligase critically regulates endoplasmic reticulum stress, ferroptosis, and metabolic homeostasis in POMC neurons. Cell Rep. 42, 112746 (2023).
Kikkert, M. et al. Human HRD1 is an E3 ubiquitin ligase involved in degradation of proteins from the endoplasmic reticulum. J. Biol. Chem. 279, 3525–3534 (2004).
Shao, S. & Hegde, R. S. Target selection during protein quality control. Trends Biochem. Sci. 41, 124–137 (2016).
Wang, S., Yang, C. I. & Shan, S. O. SecA mediates cotranslational targeting and translocation of an inner membrane protein. J. Cell Biol. 216, 3639–3653 (2017).
Perry, A. J., Hulett, J. M., Likić, V. A., Lithgow, T. & Gooley, P. R. Convergent evolution of receptors for protein import into mitochondria. Curr. Biol. 16, 221–229 (2006).
Mani, J., Meisinger, C. & Schneider, A. Peeping at TOMs—diverse entry gates to mitochondria provide insights into the evolution of eukaryotes. Mol. Biol. Evol. 33, 337–351 (2016).
He, D. et al. An alternative root for the eukaryote tree of life. Curr. Biol. 24, 465–470 (2014).
Tsaousis, A. D. et al. A functional Tom70 in the human parasite Blastocystis sp.: implications for the evolution of the mitochondrial import apparatus. Mol. Biol. Evol. 28, 781–791 (2011).
Carrie, C., Murcha, M. W. & Whelan, J. An in silico analysis of the mitochondrial protein import apparatus of plants. BMC Plant Biol. 10, 249 (2010).
Cavalier-Smith, T. Kingdoms Protozoa and Chromista and the eozoan root of the eukaryotic tree. Biol. Lett. 6, 342–345 (2010).
Gray, M. W., Burger, G. & Lang, B. F. Mitochondrial evolution. Science 283, 1476–1481 (1999).
Timmis, J. N., Ayliffe, M. A., Huang, C. Y. & Martin, W. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 5, 123–135 (2004).
Rath, S. et al. MitoCarta3.0: an updated mitochondrial proteome now with sub-organelle localization and pathway annotations. Nucleic Acids Res. 49, D1541–D1547 (2021).
Chuang, K. H., Liang, F., Higgins, R. & Wang, Y. Ubiquilin/Dsk2 promotes inclusion body formation and vacuole (lysosome)-mediated disposal of mutated huntingtin. Mol. Biol. Cell. 27, 2025–2036 (2016).
Chandel, N. S. Evolution of mitochondria as signaling organelles. Cell Metab. 22, 204–206 (2015).
Sun, Q. et al. The specific and essential role of MAVS in antiviral innate immune responses. Immunity 24, 633–642 (2006).
Vazquez, C. & Horner, S. M. MAVS coordination of antiviral innate immunity. J. Virol. 89, 6974–6977 (2015).
Meng, E. C. et al. UCSF ChimeraX: tools for structure building and analysis. Protein Sci. 32, e4792 (2023).
Hallgren, J. et al. DeepTMHMM predicts alpha and beta transmembrane proteins using deep neural networks. Preprint at bioRxiv https://doi.org/10.1101/2022.04.08.487609 (2022).
Keenan, R. J., Freymann, D. M., Stroud, R. M. & Walter, P. The signal recognition particle. Annu. Rev. Biochem. 70, 755–775 (2001).
Voorhees, R. M. & Hegde, R. S. Structures of the scanning and engaged states of the mammalian SRP–ribosome complex. eLife 4, e07975 (2015).
Mateja, A. et al. The structural basis of tail-anchored membrane protein recognition by Get3. Nature 461, 361–366 (2009).
Voorhees, R. M., Fernández, I. S., Scheres, S. H. & Hegde, R. S. Structure of the mammalian ribosome–Sec61 complex to 3.4 Å resolution. Cell 157, 1632–1643 (2014).
Schuldiner, M. et al. The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell 134, 634–645 (2008).
Miller-Vedam, L. E. et al. Structural and mechanistic basis of the EMC-dependent biogenesis of distinct transmembrane clients. eLife 9, e62611 (2020).
McGilvray, P. T. et al. An ER translocon for multi-pass membrane protein biogenesis. eLife 9, e56889 (2020).
Sundaram, A. et al. Substrate-driven assembly of a translocon for multipass membrane proteins. Nature 611, 167–172 (2022).
Acknowledgements
This work was supported by US National Institutes of Health pre-doctoral training grant T32GM007287 (to G.M.) and the Howard Hughes Medical Institute (to J.S.W.). We also thank A. Guna, K. E. Yost and J. Nunnari for careful reading of and input on the manuscript.
Author information
Authors and Affiliations
Contributions
G.M. and J.S.W. wrote the manuscript. G.M. designed the figures with the support of J.S.W.
Corresponding author
Ethics declarations
Competing interests
J.S.W. declares outside interest in 5AM Venture, Amgen, nChroma Bio, DEM BioPharma, KSQ Therapeutics, Maze Therapeutics, Tenaya Therapeutics, Tessera Therapeutics, Thermo Fisher Scientific and Third Rock Ventures. G.M. declares no competing interests.
Peer review
Peer review information
Nature Cell Biology thanks Agnieszka Chacinska, Nikolaus Pfanner and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Muthukumar, G., Weissman, J.S. Shaping the composition of the mitochondrial outer membrane. Nat Cell Biol 27, 890–901 (2025). https://doi.org/10.1038/s41556-025-01683-0
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41556-025-01683-0
This article is cited by
-
Mitochondrial fission and fusion in inflammatory diseases: mechanisms and therapeutic implications
Journal of Translational Medicine (2025)
-
Mitochondrial Calcium Channels and MAM Interaction in Calcium Homeostasis Dysregulation in Parkinson’s Disease
Neurochemical Research (2025)