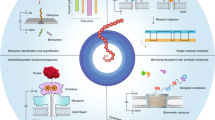
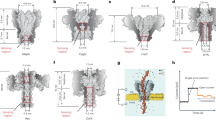
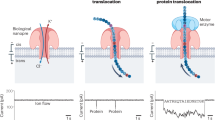
Abstract
The formation and cleavage of chemical bonds are two fundamental processes in chemistry, and the nature of these bonds determines the physical and chemical properties of a molecule. Real-time observation of chemical bonding at the single-molecule level offers insights into transient intermediates that are normally inaccessible via ensemble measurements. Protein nanopores, with their unique geometries, can be tailored into nanoreactors. Molecular bond-making and -cleavage at the reactive site of a protein nanopore’s interior wall can be visualized by monitoring ionic current changes. Therefore, nanopore-based techniques can enhance the understanding of complex binding kinetics and reaction mechanisms. In this Review we summarize recent advances in using biological nanopores as both single-molecule nanoreactors and single-molecule biosensors. The discussion covers the kinetics of single-molecule reactions under nanopore confinement, the strategies for designing biological nanopores and the latest progress in revealing reaction intermediates and pathways at the single-molecule level. Finally, we emphasize unresolved challenges and anticipate future developments in this rapidly evolving field.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Lu, H. P. Sizing up single-molecule enzymatic conformational dynamics. Chem. Soc. Rev. 43, 1118–1143 (2014).
Bayley, H., Luchian, T., Shin, S.-H. & Steffensen, M.-B. in Single Molecules and Nanotechnology (eds Rigler, R. & Vogel, H.) 251–277 (Springer, 2008).
Stone, I. et al. A single-molecule blueprint for synthesis. Nat. Rev. Chem. 5, 695–710 (2021).
Janssen, K. P. F. et al. Single molecule methods for the study of catalysis: from enzymes to heterogeneous catalysts. Chem. Soc. Rev. 43, 990–1006 (2014).
Orrit, M., Ha, T. & Sandoghdar, V. Single-molecule optical spectroscopy. Chem. Soc. Rev. 43, 973–976 (2014).
Garcia-Manyes, S. & Beedle, A. E. M. Steering chemical reactions with force. Nat. Rev. Chem. 1, 0083 (2017).
Bernardi, R. C. et al. Mechanisms of nanonewton mechanostability in a protein complex revealed by molecular dynamics simulations and single-molecule force spectroscopy. J. Am. Chem. Soc. 141, 14752–14763 (2019).
Joo, C., Balci, H., Ishitsuka, Y., Buranachai, C. & Ha, T. Advances in single-molecule fluorescence methods for molecular biology. Annu. Rev. Biochem. 77, 51–76 (2008).
Kasianowicz, J., Walker, B., Krishnasastry, M. & Bayley, H. Genetically engineered pores as metal ion biosensors. Mat. Res. Soc. Symp. Proc. 330, 217–223 (1993).
Mindell, J. A., Zhan, H., Huynh, P. D., Collier, R. J. & Finkelstein, A. Reaction of diphtheria toxin channels with sulfhydryl-specific reagents: observation of chemical reactions at the single molecule level. Proc. Natl Acad. Sci. USA 91, 5272–5276 (1994).
Walker, B., Kasianowicz, J., Krishnasastry, M. & Bayley, H. A pore-forming protein with a metal-actuated switch. Protein Eng. Des. Sel. 7, 655–662 (1994).
Kasianowicz, J. J., Brandin, E., Branton, D. & Deamer, D. W. Characterization of individual polynucleotide molecules using a membrane channel. Proc. Natl Acad. Sci. USA 93, 13770–13773 (1996).
Braha, O. et al. Designed protein pores as components for biosensors. Chem. Biol. 4, 497–505 (1997).
Kasianowicz, J. J., Burden, D. L., Han, L. C., Cheley, S. & Bayley, H. Genetically engineered metal ion binding sites on the outside of a channel’s transmembrane β-barrel. Biophys. J. 76, 837–845 (1999).
Guan, X., Gu, L.-Q., Cheley, S., Braha, O. & Bayley, H. Stochastic sensing of TNT with a genetically engineered pore. ChemBioChem 6, 1875–1881 (2005).
Branton, D. et al. The potential and challenges of nanopore sequencing. Nat. Biotechnol. 26, 1146–1153 (2008).
Meller, A., Nivon, L., Brandin, E., Golovchenko, J. & Branton, D. Rapid nanopore discrimination between single polynucleotide molecules. Proc. Natl Acad. Sci. USA 97, 1079–1084 (2000).
Meller, A., Nivon, L. & Branton, D. Voltage-driven DNA translocations through a nanopore. Phys. Rev. Lett. 86, 3435–3438 (2001).
Schreiber, J. et al. Error rates for nanopore discrimination among cytosine, methylcytosine, and hydroxymethylcytosine along individual DNA strands. Proc. Natl Acad. Sci. USA 110, 18910–18915 (2013).
Sutherland, T. C. et al. Structure of peptides investigated by nanopore analysis. Nano Lett. 4, 1273–1277 (2004).
Movileanu, L., Schmittschmitt, J. P., Scholtz, J. M. & Bayley, H. Interactions of peptides with a protein pore. Biophys. J. 89, 1030–1045 (2005).
Piguet, F. et al. Identification of single amino acid differences in uniformly charged homopolymeric peptides with aerolysin nanopore. Nat. Commun. 9, 966 (2018).
Shimizu, K. et al. De novo design of a nanopore for single-molecule detection that incorporates a β-hairpin peptide. Nat. Nanotechnol. 17, 67–75 (2022).
Pastoriza-Gallego, M. et al. Dynamics of unfolded protein transport through an aerolysin pore. J. Am. Chem. Soc. 133, 2923–2931 (2011).
Thakur, A. K. & Movileanu, L. Single-molecule protein detection in a biofluid using a quantitative nanopore sensor. ACS Sens. 4, 2320–2326 (2019).
Sauciuc, A., Morozzo Della Rocca, B., Tadema, M. J., Chinappi, M. & Maglia, G. Translocation of linearized full-length proteins through an engineered nanopore under opposing electrophoretic force. Nat. Biotechnol. 42, 1275–1281 (2024).
Shin, S.-H., Steffensen, M. B., Claridge, T. D. W. & Bayley, H. Formation of a chiral center and pyrimidal inversion at the single‐molecule level. Angew. Chem. Int. Ed. 46, 7412–7416 (2007).
Pulcu, G. S., Mikhailova, E., Choi, L.-S. & Bayley, H. Continuous observation of the stochastic motion of an individual small-molecule walker. Nat. Nanotechol. 10, 76–83 (2015).
Hornblower, B. et al. Single-molecule analysis of DNA–protein complexes using nanopores. Nat. Methods 4, 315–317 (2007).
Li, T., Liu, L., Li, Y., Xie, J. & Wu, H.-C. A universal strategy for aptamer‐based nanopore sensing through host–guest interactions inside α-hemolysin. Angew. Chem. Int. Ed. 54, 7568–7571 (2015).
Jeong, K.-B. et al. Single-molecule fingerprinting of protein–drug interaction using a funneled biological nanopore. Nat. Commun. 14, 1461 (2023).
Manrao, E. A. et al. Reading DNA at single-nucleotide resolution with a mutant MspA nanopore and phi29 DNA polymerase. Nat. Biotechnol. 30, 349–353 (2012).
Deamer, D., Akeson, M. & Branton, D. Three decades of nanopore sequencing. Nat. Biotechnol. 34, 518–524 (2016).
Ying, Y.-L. et al. Nanopore-based technologies beyond DNA sequencing. Nat. Nanotechnol. 17, 1136–1146 (2022).
Jiang, J. et al. Protein nanopore reveals the renin–angiotensin system crosstalk with single-amino-acid resolution. Nat. Chem. 15, 578–586 (2023).
Koch, C. et al. Nanopore sequencing of DNA-barcoded probes for highly multiplexed detection of microRNA, proteins and small biomarkers. Nat. Nanotechnol. 18, 1483–1491 (2023).
Ying, Y.-L. & Long, Y.-T. Nanopore-based single-biomolecule interfaces: from information to knowledge. J. Am. Chem. Soc. 141, 15720–15729 (2019).
Thakur, A. K. & Movileanu, L. Real-time measurement of protein–protein interactions at single-molecule resolution using a biological nanopore. Nat. Biotechnol. 37, 96–101 (2019).
Kawano, R. et al. Rapid detection of a cocaine-binding aptamer using biological nanopores on a chip. J. Am. Chem. Soc. 133, 8474–8477 (2011).
Ma, W. et al. Nanopore electrochemical sensors for emerging hazardous pollutants detection. Electrochim. Acta 475, 143678 (2024).
Luchian, T., Shin, S.-H. & Bayley, H. Kinetics of a three‐step reaction observed at the single‐molecule level. Angew. Chem. Int. Ed. 42, 1926–1929 (2003).
Lu, S., Li, W.-W., Rotem, D., Mikhailova, E. & Bayley, H. A primary hydrogen–deuterium isotope effect observed at the single-molecule level. Nat. Chem. 2, 921–928 (2010).
Lee, J. et al. Semisynthetic nanoreactor for reversible single-molecule covalent chemistry. ACS Nano 10, 8843–8850 (2016).
Zhou, B., Wang, Y.-Q., Cao, C., Li, D.-W. & Long, Y.-T. Monitoring disulfide bonds making and breaking in biological nanopore at single molecule level. Sci. China Chem. 61, 1385–1388 (2018).
Qiu, K., Fato, T. P., Yuan, B. & Long, Y.-T. Toward precision measurement and manipulation of single‐molecule reactions by a confined space. Small 15, 1805426 (2019).
Dong, B. et al. Deciphering nanoconfinement effects on molecular orientation and reaction intermediate by single molecule imaging. Nat. Commun. 10, 4815 (2019).
Galenkamp, N. S., Biesemans, A. & Maglia, G. Directional conformer exchange in dihydrofolate reductase revealed by single-molecule nanopore recordings. Nat. Chem. 12, 481–488 (2020).
Jia, W. et al. Programmable nano-reactors for stochastic sensing. Nat. Commun. 12, 5811 (2021).
Soskine, M. et al. An engineered ClyA nanopore detects folded target proteins by selective external association and pore entry. Nano Lett. 12, 4895–4900 (2012).
Lee, J. & Bayley, H. Semisynthetic protein nanoreactor for single-molecule chemistry. Proc. Natl Acad. Sci. USA 112, 13768–13773 (2015).
Qing, Y., Ionescu, S. A., Pulcu, G. S. & Bayley, H. Directional control of a processive molecular hopper. Science 361, 908–912 (2018).
Qing, Y., Pulcu, G. S., Bell, N. A. W. & Bayley, H. Bioorthogonal cycloadditions with sub-millisecond intermediates. Angew. Chem. Int. Ed. 57, 1218–1221 (2018).
Haugland, M. M., Borsley, S., Cairns-Gibson, D. F., Elmi, A. & Cockroft, S. L. Synthetically diversified protein nanopores: resolving click reaction mechanisms. ACS Nano 13, 4101–4110 (2019).
Gu, L.-Q., Braha, O., Conlan, S., Cheley, S. & Bayley, H. Stochastic sensing of organic analytes by a pore-forming protein containing a molecular adapter. Nature 398, 686–690 (1999).
Ivica, J., Williamson, P. T. F. & De Planque, M. R. R. Salt gradient modulation of microRNA translocation through a biological nanopore. Anal. Chem. 89, 8822–8829 (2017).
Brun, L. et al. Dynamics of polyelectrolyte transport through a protein channel as a function of applied voltage. Phys. Rev. Lett. 100, 158302 (2008).
Piguet, F. et al. Electroosmosis through α-hemolysin that depends on alkali cation type. J. Phys. Chem. Lett. 5, 4362–4367 (2014).
Huang, G., Willems, K., Soskine, M., Wloka, C. & Maglia, G. Electro-osmotic capture and ionic discrimination of peptide and protein biomarkers with FraC nanopores. Nat. Commun. 8, 935 (2017).
Gu, L.-Q., Cheley, S. & Bayley, H. Electroosmotic enhancement of the binding of a neutral molecule to a transmembrane pore. Proc. Natl Acad. Sci. USA 100, 15498–15503 (2003).
Boukhet, M. et al. Probing driving forces in aerolysin and α-hemolysin biological nanopores: electrophoresis versus electroosmosis. Nanoscale 8, 18352–18359 (2016).
Asandei, A. et al. Electroosmotic trap against the electrophoretic force near a protein nanopore reveals peptide dynamics during capture and translocation. ACS Appl. Mater. Interfaces 8, 13166–13179 (2016).
Liu, W., Yang, Z.-L., Yang, C.-N., Ying, Y.-L. & Long, Y.-T. Profiling single-molecule reaction kinetics under nanopore confinement. Chem. Sci. 13, 4109–4114 (2022).
Luchian, T., Shin, S.-H. & Bayley, H. Single‐molecule covalent chemistry with spatially separated reactants. Angew. Chem. Int. Ed. 42, 3766–3771 (2003).
Yang, C.-N. et al. Electrochemical visualization of single-molecule thiol substitution with nanopore measurement. ACS Meas. Sci. Au 4, 76–80 (2024).
Ramsay, W. J., Bell, N. A. W., Qing, Y. & Bayley, H. Single-molecule observation of the intermediates in a catalytic cycle. J. Am. Chem. Soc. 140, 17538–17546 (2018).
Cao, J. et al. Giant single molecule chemistry events observed from a tetrachloroaurate(III) embedded Mycobacterium smegmatis porin A nanopore. Nat. Commun. 10, 5668 (2019).
Yang, C.-N. et al. Electrochemical kinetic fingerprinting of single-molecule cooridations in the confined nanopores. Faraday Discuss. 257, 29–43 (2025).
Zhang, S. et al. A nanopore‐based saccharide sensor. Angew. Chem. Int. Ed. 61, e202203769 (2022).
Wu, X.-Y. et al. Controlled genetic encoding of unnatural amino acids in a protein nanopore. Angew. Chem. Int. Ed. 62, e202300582 (2023).
Yang, J. et al. Site‐specific introduction of bioorthogonal handles to nanopores by genetic code expansion. Angew. Chem. Int. Ed. 62, e202216115 (2023).
Jiang, X. et al. Single-molecule observation of selenoenzyme intermediates in a semisynthetic seleno-α-hemolysin nanoreactor. Anal. Chem. 94, 8433–8440 (2022).
Niu, H., Li, M.-Y., Ying, Y.-L. & Long, Y.-T. An engineered third electrostatic constriction of aerolysin to manipulate heterogeneously charged peptide transport. Chem. Sci. 13, 2456–2461 (2022).
Beckstein, O., Tai, K. & Sansom, M. S. P. Not ions alone: barriers to ion permeation in nanopores and channels. J. Am. Chem. Soc. 126, 14694–14695 (2004).
Willems, K. et al. Engineering and modeling the electrophoretic trapping of a single protein inside a nanopore. ACS Nano 13, 9980–9992 (2019).
Wang, Y.-Q. et al. Rationally designed sensing selectivity and sensitivity of an aerolysin nanopore via site-directed mutagenesis. ACS Sens. 3, 779–783 (2018).
Soskine, M., Biesemans, A., De Maeyer, M. & Maglia, G. Tuning the size and properties of ClyA nanopores assisted by directed evolution. J. Am. Chem. Soc. 135, 13456–13463 (2013).
Wang, Y.-Q. et al. Identification of essential sensitive regions of the aerolysin nanopore for single oligonucleotide analysis. Anal. Chem. 90, 7790–7794 (2018).
Liu, W. et al. Observing confined local oxygen‐induced reversible thiol/disulfide cycle with a protein nanopore. Angew. Chem. Int. Ed. 135, e202304023 (2023).
Lucas, F. L. R. et al. The manipulation of the internal hydrophobicity of FraC nanopores augments peptide capture and recognition. ACS Nano 15, 9600–9613 (2021).
Rosenstein, J. K., Wanunu, M., Merchant, C. A., Drndic, M. & Shepard, K. L. Integrated nanopore sensing platform with sub-microsecond temporal resolution. Nat. Methods 9, 487–492 (2012).
Shin, S.-H., Luchian, T., Cheley, S., Braha, O. & Bayley, H. Kinetics of a reversible covalent-bond-forming reaction observed at the single-molecule level. Angew. Chem. Int. Ed. 41, 3707–3709 (2002).
Steffensen, M. B., Rotem, D. & Bayley, H. Single-molecule analysis of chirality in a multicomponent reaction network. Nat. Chem. 6, 603–607 (2014).
Shin, S.-H. & Bayley, H. Stepwise growth of a single polymer chain. J. Am. Chem. Soc. 127, 10462–10463 (2005).
Qing, Y., Tamagaki-Asahina, H., Ionescu, S. A., Liu, M. D. & Bayley, H. Catalytic site-selective substrate processing within a tubular nanoreactor. Nat. Nanotechnol. 14, 1135–1142 (2019).
Qing, Y. & Bayley, H. Enzymeless DNA base identification by chemical stepping in a nanopore. J. Am. Chem. Soc. 143, 18181–18187 (2021).
Liu, W. et al. Single-molecule sensing inside stereo- and regio-defined hetero-nanopores. Nat. Nanotechnol. 19, 1693–1701 (2024).
Rieth, A. J., Wright, A. M. & Dincă, M. Kinetic stability of metal–organic frameworks for corrosive and coordinating gas capture. Nat. Rev. Mater. 4, 708–725 (2019).
Qian, Q. et al. MOF-based membranes for gas separations. Chem. Rev. 120, 8161–8266 (2020).
Allendorf, M. D. et al. Challenges to developing materials for the transport and storage of hydrogen. Nat. Chem. 14, 1214–1223 (2022).
Bo, Z., Lim, Z. H., Duarte, F., Bayley, H. & Qing, Y. Mobile molecules: reactivity profiling guides faster movement on a cysteine track. Angew. Chem. Int. Ed. 135, e202300890 (2023).
Berezin, M. Y. & Achilefu, S. Fluorescence lifetime measurements and biological imaging. Chem. Rev. 110, 2641–2684 (2010).
Roithová, J. Characterization of reaction intermediates by ion spectroscopy. Chem. Soc. Rev. 41, 547–559 (2012).
Lin, J. J.-M. & Chao, W. Structure-dependent reactivity of Criegee intermediates studied with spectroscopic methods. Chem. Soc. Rev. 46, 7483–7497 (2017).
Wu, Y.-J., Takahashi, K. & Lin, J. J.-M. Kinetics of the simplest Criegee intermediate reaction with water vapor: revisit and isotope effect. J. Phys. Chem. A 127, 8059–8072 (2023).
Li, X. et al. Emerging data processing methods for single‐entity electrochemistry. Angew. Chem. Int. Ed. 136, e202316551 (2024).
Liu, L., Yang, C., Zhao, K., Li, J. & Wu, H.-C. Ultrashort single-walled carbon nanotubes in a lipid bilayer as a new nanopore sensor. Nat. Commun. 4, 2989 (2013).
Geng, J. et al. Stochastic transport through carbon nanotubes in lipid bilayers and live cell membranes. Nature 514, 612–615 (2014).
Feng, J. et al. Observation of ionic Coulomb blockade in nanopores. Nat. Mater. 15, 850–855 (2016).
Tunuguntla, R. H. et al. Enhanced water permeability and tunable ion selectivity in subnanometer carbon nanotube porins. Science 357, 792–796 (2017).
Wang, M. et al. Dynamic curvature nanochannel‐based membrane with anomalous ionic transport behaviors and reversible rectification switch. Adv. Mater. 31, 1805130 (2019).
Huang, S., Romero-Ruiz, M., Castell, O. K., Bayley, H. & Wallace, M. I. High-throughput optical sensing of nucleic acids in a nanopore array. Nat. Nanotechnol. 10, 986–991 (2015).
Weiss, M. et al. Sequential bottom-up assembly of mechanically stabilized synthetic cells by microfluidics. Nat. Mater. 17, 89–96 (2018).
Zhang, Y. et al. A microscale soft ionic power source modulates neuronal network activity. Nature 620, 1001–1006 (2023).
Acknowledgements
This research was supported by the National Key R&D Program of China (2022YFA1304604), the National Natural Science Foundation of China (2247040607, 22027806 and 22334006) and programs for high-level entrepreneurial and innovative talent introduction of Jiangsu Province.
Author information
Authors and Affiliations
Contributions
Y.-L.Y. and Y.-T.L. conceived the concept of this Review. Y.-L.Y., C.-N.Y., W.L. and X.-Y.W. wrote the manuscript. C.-N.Y., W.L. and Y.-L.Y. prepared the figures. All authors contributed to the reviewing and editing of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Chemistry thanks Manoj Varma, Xu Hou and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ying, YL., Yang, CN., Liu, W. et al. Understanding single-molecule reactions using nanopore-based techniques. Nat. Chem. 17, 1450–1461 (2025). https://doi.org/10.1038/s41557-025-01905-w
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41557-025-01905-w