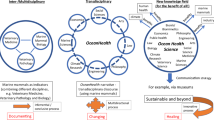
Abstract
Reproducibility has long been a tenet of science but has been challenging to achieve—we learned this the hard way when our old approaches proved inadequate to efficiently reproduce our own work. Here we describe how several free software tools have fundamentally upgraded our approach to collaborative research, making our entire workflow more transparent and streamlined. By describing specific tools and how we incrementally began using them for the Ocean Health Index project, we hope to encourage others in the scientific community to do the same—so we can all produce better science in less time.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Baker, M. Over half of psychology studies fail reproducibility test. Nature Newshttp://dx.doi.org/10.1038/nature.2015.18248 (2015).
Baker, M. & Dolgin, E. Cancer reproducibility project releases first results. Nature Newshttp://dx.doi.org/10.1038/541269a (2017).
Open Science Collaboration. Estimating the reproducibility of psychological science. Science 349, aac4716 (2015).
Baker, M. 1,500 scientists lift the lid on reproducibility. Nature 533, 452–454 (2016).
Aschwanden, C. Science isn't broken. FiveThirtyEighthttp://go.nature.com/2qaz0Tz (19 August 2015)
Buck, S. Solving reproducibility. Science 348, 1403–1403 (2015).
Frew, J. & Dozier, J. Environmental informatics. Annu. Rev. Environ. Resources 37, 449–472 (2012).
Jones, M. B., Schildhauer, M. P., Reichman, O. J. & Bowers, S. The new bioinformatics: Integrating ecological data from the gene to the biosphere. Annu. Rev. Ecol. Evol. Syst. 37, 519–544 (2006).
Michener, W. K. & Jones, M. B. Ecoinformatics: Supporting ecology as a data-intensive science. Trends Ecol. Evol. 27, 85–93 (2012).
Check Hayden, E. Mozilla plan seeks to debug scientific code. Nature Newshttp://dx.doi.org/10.1038/501472a (2013).
Boettiger, C., Chamberlain, S., Hart, E. & Ram, K. Building software, building community: Lessons from the rOpenSci project. J. Open Res. Softw. 3, e8 (2015).
Wilson, G. et al. Good enough practices in scientific computing. Preprint at https://arxiv.org/abs/1609.00037 (2016).
Wilson, G. V. Where's the real bottleneck in scientific computing? Am. Sci. 94, 5–6 (2006).
Baker, M. Scientific computing: Code alert. Nature 541, 563–565 (2017).
Barone, L., Williams, J. & Micklos, D. Unmet needs for analyzing biological big data: A survey of 704 NSF principal investigators. Preprint at bioRxivhttps://doi.org/10.1101/108555 (2017).
Wolkovich, E. M., Regetz, J. & O’Connor, M. I. Advances in global change research require open science by individual researchers. Global Change Biol. 18, 2102–2110 (2012).
Nosek, B. A. et al. Promoting an open research culture. Science 348, 1422–1425 (2015).
Reichman, O. J., Jones, M. B. & Schildhauer, M. P. Challenges and opportunities of open data in ecology. Science 331, 703–705 (2011).
Shade, A. & Teal, T. K. Computing workflows for biologists: A roadmap. PLoS Biol. 13, e1002303 (2015).
Goodman, A. et al. Ten simple rules for the care and feeding of scientific data. PLoS Comput. Biol. 10, e1003542 (2014).
Sandve, G. K., Nekrutenko, A., Taylor, J. & Hovig, E. Ten simple rules for reproducible computational research. PLoS Comput. Biol. 9, e1003285 (2013).
White, E. P. et al. Nine simple ways to make it easier to (re)use your data. Ideas Ecol. Evol.http://doi.org/10.4033/iee.2013.6b.6.f (2013).
Kervin, K., Michener, W. & Cook, R. Common errors in ecological data sharing. J. eScience Librarianshiphttp://dx.doi.org/10.7191/jeslib.2013.1024 (2013).
Lewandowsky, S. & Bishop, D. Research integrity: don't let transparency damage science. Nature Newshttp://dx.doi.org/10.1038/529459a (2016).
Michener, W. K. Ten simple rules for creating a good data management plan. PLoS Comput. Biol. 11, e1004525 (2015).
Mislan, K. A. S., Heer, J. M. & White, E. P. Elevating the status of code in ecology. Trends Ecol. Evol. 31, 4–7 (2016).
Kratz, J. & Strasser, C. Data publication consensus and controversies. F1000Researchhttp://dx.doi.org/10.12688/f1000research.3979.3 (2014).
Munafò, M. R. et al. A manifesto for reproducible science. Nat. Hum. Behav. 1, 0021 (2017).
Martinez, C. et al. Reproducibility in Science: A Guide to Enhancing Reproducibility in Scientific Results and Writinghttp://ropensci.github.io/reproducibility-guide/ (2014).
Tuyl, S. V. & Whitmire, A. L. Water, water, everywhere: defining and assessing data sharing in academia. PLoS ONE 11, e0147942 (2016).
Baker, M. Why scientists must share their research code. Nature Newshttp://dx.doi.org/10.1038/nature.2016.20504 (2016).
Kidwell, M. C. et al. Badges to acknowledge open practices: A simple, low-cost, effective method for increasing transparency. PLoS Biol. 14, e1002456 (2016).
Wickham, H. & Grolemund, G. R for Data Science (O’Reilly, 2016); http://r4ds.had.co.nz/
Lowndes, J. S. S. et al. Best practices for assessing ocean health in multiple contexts using tailorable frameworks. PeerJ 3, e1503 (2015).
Lowndes, J. A biography of the ocean health index. ohi-sciencehttp://ohi-science.org/news/Biography-OHI (13 January 2017).
Halpern, B. S. et al. An index to assess the health and benefits of the global ocean. Nature 488, 615–620 (2012).
Halpern, B. S. et al. Patterns and emerging trends in global ocean health. PLoS ONE 10, e0117863 (2015).
Five years of global ocean health index assessments. ohi-sciencehttp://ohi-science.org/ohi-global (2016).
Goldfuss, C. & Holdren, J. P. The nation's first ocean plans. The White Househttps://obamawhitehouse.archives.gov/blog/2016/12/07/nations-first-ocean-plans (7 December 2016).
Hampton, S. E. et al. The tao of open science for ecology. Ecosphere 6, art 120 (2015).
Mills, B. Introducing mozilla science study groups. Mozillahttps://science.mozilla.org/blog/introducing-mozilla-science-study-groups (22 April 2015).
R Core Team R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2016); https://www.R-project.org/
RStudio Team RStudio: Integrated Development for R (RStudio, 2016); www.rstudio.com
Git Team Git Version Control System (Git, 2016); https://git-scm.com/
GitHub: A Collaborative Online Platform To Build Software (GitHub, 2016); https://github.com
Wilson, G. V. Software carpentry: getting scientists to write better code by making them more productive. Comput. Sci. Eng. 8, 66–69 (2006).
Broman, K. Initial steps toward reproducible research. http://kbroman.org/steps2rr/ (2016).
McKiernan, E. C. et al. How open science helps researchers succeed. eLife 5, e16800 (2016).
Seltenrich, N. Scaling the heights of data science. Breakthroughshttps://nature.berkeley.edu/breakthroughs/opensci-data (2016).
Wilson, G. et al. Best practices for scientific computing. PLoS Biol. 12, e1001745 (2014).
Haddock, S. H. & Dunn, C. W. Practical Computing for Biologists (Sinauer Associates, 2011).
Barnes, N. Publish your computer code: it is good enough. Nature 467, 753 (2010).
Frazier, M., Longo, C. & Halpern, B. S. Mapping uncertainty due to missing data in the global ocean health index. PLoS ONE 11, e0160377 (2016).
ESRI ArcGIS Platform (2016); http://www.esri.com/software/arcgis
The QGIS Team QGIS Project (2016); http://www.qgis.org
The Python Team Python (2016); https://www.python.org/
Wickham, H. Tidy data. J. Stat. Softw. 59, 1–23 (2014).
Wickham, H. Tidyverse Tidyweb (accessed 29 January 2017); http://tidyverse.org/
Wickham, H. Tidyverse: Easily Install and Load ‘tidyverse’ Packages (2016); https://CRAN.R-project.org/package=tidyverse
Fischetti, T. How dplyr replaced my most common r idioms. StatsBlogshttp://www.onthelambda.com/2014/02/10/how-dplyr-replaced-my-most-common-r-idioms/ (10 February 2014).
RStudio Team R Markdown (2016); http://rmarkdown.rstudio.com/
Allaire, J. J. et al. R Markdown: Dynamic Documents for R (2016); https://CRAN.R-project.org/package=rmarkdown
Ocean Health Index ohicore Package (Ocean Health Index Team, 2016).
Wickham, H. R Packages (O’Reilly, 2015); http://r-pkgs.had.co.nz/
Wickham, H. & Chang, W. Devtools: Tools to Make Developing R Packages Easier (2016); https://CRAN.R-project.org/package=devtools
Wickham, H., Danenberg, P. & Eugster, M. Roxygen2: In-Source Documentation for R (2015); https://CRAN.R-project.org/package=roxygen2
Ram, K. Git can facilitate greater reproducibility and increased transparency in science. Source Code Biol. Med. 8, 7 (2013).
Blischak, J. D., Davenport, E. R. & Wilson, G. A quick introduction to version control with git and GitHub. PLoS Comput. Biol. 12, e1004668 (2016).
Perez-Riverol, Y. et al. Ten simple rules for taking advantage of Git and GitHub. PLoS Comput. Biol. 12, e1004947 (2016).
Duhigg, C. What Google learned from its quest to build the perfect team. The New York Times (25 February 2016).
Perkel, J. Democratic databases: Science on GitHub. Nature 538, 127–128 (2016).
Casadevall, A. & Fang, F. C. Reproducible science. Infect. Immun. 78, 4972–4975 (2010).
Wilson, G. Software carpentry: lessons learned. F1000Researchhttp://dx.doi.org/10.12688/f1000research.3-62.v2 (2016).
Hampton, S. E. et al. Big data and the future of ecology. Front. Ecol. Environ. 11, 156–162 (2013).
Lohr, S. For big-data scientists, ‘janitor work’ is key hurdle to insights. The New York Times (17 August 2014).
FitzJohn, R., Pennell, M., Zanne, A. & Cornell, W. Reproducible research is still a challenge. ROpenSci https://ropensci.org//blog/2014/06/09/reproducibility/ (9 June 2014).
Boland, M. R., Karczewski, K. J. & Tatonetti, N. P. Ten simple rules to enable multi-site collaborations through data sharing. PLoS Comput. Biol. 13, e1005278 (2017).
Perkel, J. M. Scientific writing: the online cooperative. Nature 514, 127–128 (2014).
How Twitter improved my ecological model. R-bloggershttps://www.r-bloggers.com/how-twitter-improved-my-ecological-model/ (26 February 2015).
Acknowledgements
The Ocean Health Index is a collaboration between Conservation International and the National Center for Ecological Analysis and Synthesis at the University of California at Santa Barbara. We thank J. Polsenberg, S. Katona, E. Pacheco and L. Mosher who are our partners at Conservation International. We thank all past contributors and funders that have supported the Ocean Health Index, including B. Wrigley and H. Wrigley and The Pacific Life Foundation. We also thank all the individuals and groups that openly make their data, tools and tutorials freely available to others. Finally, we thank H. Wickham, K. Ram, K. Woo and M. Schildhauer for friendly review of the developing manuscript. See http://ohi-science.org/betterscienceinlesstime as an example of a website built with RMarkdown and the RStudio–GitHub workflow, and for links and resources referenced in the paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Lowndes, J., Best, B., Scarborough, C. et al. Our path to better science in less time using open data science tools. Nat Ecol Evol 1, 0160 (2017). https://doi.org/10.1038/s41559-017-0160
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41559-017-0160
This article is cited by
-
Transparency in epidemiological analyses of cohort data a case study of the Norwegian mother, father, and child cohort study (MoBa)
BMC Medical Research Methodology (2025)
-
Innovative and practical tools for monitoring and assessing biodiversity status and impacts of multiple human pressures in marine systems
Environmental Monitoring and Assessment (2024)
-
The challenges of research data management in cardiovascular science: a DGK and DZHK position paper—executive summary
Clinical Research in Cardiology (2024)
-
Protocol for developing a dashboard for interactive cohort analysis of oral health-related data
BMC Oral Health (2023)
-
Incentivising open ecological data using blockchain technology
Scientific Data (2023)