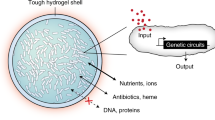
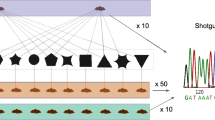
Abstract
Emerging products of biotechnology involve the release of living genetically modified microbes (GMMs) into the environment. However, regulatory challenges limit their use. So far, GMMs have mainly been tested in agriculture and environmental cleanup, with few approved for commercial purposes. Current government regulations do not sufficiently address modern genetic engineering and limit the potential of new applications, including living therapeutics, engineered living materials, self-healing infrastructure, anticorrosion coatings and consumer products. Here, based on 47 global studies on soil-released GMMs and laboratory microcosm experiments, we discuss the environmental behaviour of released bacteria and offer engineering strategies to help improve performance, control persistence and reduce risk. Furthermore, advanced technologies that improve GMM function and control, but lead to increases in regulatory scrutiny, are reviewed. Finally, we propose a new regulatory framework informed by recent data to maximize the benefits of GMMs and address risks.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Change history
24 February 2025
A Correction to this paper has been published: https://doi.org/10.1038/s41564-025-01956-2
References
Velkov, V. V. Environmental genetic engineering: hope or hazard? Curr. Sci. 70, 823–832 (1996).
Waltz, E. Small innovators advance microbes as alternatives to chemical crop sprays. Nat. Biotechnol. 41, 162–164 (2023).
Voigt, C. A. Synthetic biology 2020–2030: six commercially-available products that are changing our world. Nat. Commun. 11, 6379 (2020).
Gallup, O., Ming, H. & Ellis, T. Ten future challenges for synthetic biology. Eng. Biol. 5, 51–59 (2021).
Brooks, S. M. & Alper, H. S. Applications, challenges and needs for employing synthetic biology beyond the lab. Nat. Commun. 12, 1390 (2021).
Berg, P. & Singer, M. F. The recombinant DNA controversy: twenty years later. Proc. Natl Acad. Sci. USA 92, 9011–9013 (1995).
Goeddel, D. V. et al. Direct expression in Escherichia coli of a DNA sequence coding for human growth hormone. Nature 281, 544–548 (1979).
Mbanya, J. C., Sandow, J., Landgraf, W. & Owens, D. R. Recombinant human insulin in global diabetes management–focus on clinical efficacy. Eur. Endocrinol. 13, 21–25 (2017).
Chakrabarty, A. M. Microorganisms having multiple compatible degradative energy-generating plasmids and preparation thereof. US patent US4,259,444A (1972).
Wrubel, R. P., Krimsky, S. & Anderson, M. D. Regulatory oversight of genetically engineered microorganisms: has regulation inhibited innovation? Environ. Manag. 21, 571–586 (1997).
Carter, S. R., Rodemeyer, M., Garfinkel, M. S. & Friedman, R. M. Synthetic Biology and the US Biotechnology Regulatory System: Challenges and Options (J. Craig Venter Institute, 2014).
National Academies of Sciences, Engineering & Medicine. in Preparing for Future Products of Biotechnology 72 (National Academies Press, 2017).
Environmental Protection Agency—Office of Pollution Prevention and Toxics. Points to Consider in the Preparation of TSCA Biotechnology Submissions for Microorganisms (EPA, 1997); https://www.epa.gov/sites/default/files/2015-08/documents/biotech_points_to_consider.pdf
Environmental Protection Agency. in Pesticide Registration Manual Ch. 3 (EPA, 2024); https://www.epa.gov/pesticide-registration/pesticide-registration-manual-chapter-3-additional-considerations
US Government Biotechnology Working Group. Modernizing the Regulatory System for Biotechnology Products: Final Version of the 2017 Update to the Coordinated Framework for the Regulation of Biotechnology (EPA, 2017); https://www.epa.gov/sites/default/files/2017-01/documents/2017_coordinated_framework_update.pdf
Coordinated Framework for Regulation of Biotechnology. Report No. FR 23302 (Office of Science and Technology Policy, 1986).
Wozniak, C. A., McClung, G., Gagliardi, J., Segal, M. & Matthews, K. in Regulation of Agricultural Biotechnology: the United States and Canada (eds Wozniak, C. A. & McHughen, A.) Ch. 4, 57–94 (Springer, 2012); https://doi.org/10.1007/978-94-007-2156-2_4 (2012).
Microbial Pesticides Data Requirements. Code of Federal Regulations (United States Government Publishing Office, 2020); https://www.govinfo.gov/app/details/CFR-2020-title40-vol26/CFR-2020-title40-vol26-sec158-2110
US Department of Agriculture, Animal and Plant Health Inspection Service. Guide for Submitting Permit Applications for Microorganisms Developed Using Genetic Engineering Under 7 CFR part 340 (US Department of Agriculture, 2023); https://www.aphis.usda.gov/biotechnology/downloads/draft-brs-microbe-permit-guide.pdf
US Department of Agriculture, Animal and Plant Inspection Service. Laws and Regulations (US Department of Agriculture, 2024); https://www.aphis.usda.gov/laws-regs
Council on Environmental Quality Executive Office of the President. A Citizen’s Guide to the NEPA. Having Your Voice Heard (US Department of Envornment, 2007); https://ceq.doe.gov/docs/get-involved/Citizens_Guide_Dec07.pdf
US Food and Drug Administration. What We Do (ES EPA, 2023); https://www.fda.gov/about-fda/what-we-do#:~:text=Information%20for%20Consumers-,FDA%20Mission,and%20products%20that%20emit%20radiation
Hoffman, N. E. Revisions to USDA biotechnology regulations: the SECURE rule. Proc. Natl Acad. Sci. USA 118, e2004841118 (2021).
Graham, A. E. & Ledesma-Amaro, R. The microbial food revolution. Nat. Commun. 14, 2231 (2023).
US Food & Drug Administration. Center for Biologics Evaluation and Research (CBER) (US FDA, 2024); https://www.fda.gov/about-fda/fda-organization/center-biologics-evaluation-and-research-cber
US Food & Drug Administration. Generally Recognized as Safe (GRAS) (US FDA, 2023); https://www.fda.gov/food/food-ingredients-packaging/generally-recognized-safe-gras
McCold, L. N. & Saulsbury, J. W. Defining the no-action alternative for national environmental policy act analyses of continuing actions. Environ. Impact Assess. Rev. 18, 15–37 (1998).
Wang, W. et al. Harnessing the hygroscopic and biofluorescent behaviors of genetically tractable microbial cells to design biohybrid wearables. Sci. Adv. 3, e1601984 (2017).
Manfredini, A. et al. Current methods, common practices, and perspectives in tracking and monitoring bioinoculants in soil. Front. Microbiol. 12, 698491 (2021).
Van Elsas, J., Duarte, G., Rosado, A. & Smalla, K. Microbiological and molecular biological methods for monitoring microbial inoculants and their effects in the soil environment. J. Microbiol. Methods 32, 133–154 (1998).
Corich, V. et al. Aspects of marker/reporter stability and selectivity in soil microbiology. Microb. Ecol. 41, 333–340 (2001).
Rastogi, G., Tech, J. J., Coaker, G. L. & Leveau, J. H. J. A PCR-based toolbox for the culture-independent quantification of total bacterial abundances in plant environments. J. Microbiol. Methods 83, 127–132 (2010).
Techtmann, S. M. & Hazen, T. C. Metagenomic applications in environmental monitoring and bioremediation. J. Ind. Microbiol. Biotechnol. 43, 1345–1354 (2016).
Shen, J., McFarland, A. G., Young, V. B., Hayden, M. K. & Hartmann, E. M. Toward accurate and robust environmental surveillance using metagenomics. Front. Genet. 12, 600111 (2021).
Recorbet, G. et al. Conditional suicide system of Escherichia coli released into soil that uses the Bacillus subtilis sacB gene. Appl. Environ. Microbiol. 59, 1361–1366 (1993).
Scanferlato, V. S., Orvos, D. R., Cairns, J. & Lacy, G. H. Genetically engineered Erwinia carotovora in aquatic microcosms: survival and effects on functional groups of indigenous bacteria. Appl. Environ. Microbiol. 55, 1477–1482 (1989).
Franz, E. et al. Manure-amended soil characteristics affecting the survival of E. coli O157:H7 in 36 Dutch soils. Environ. Microbiol. 10, 313–327 (2008).
Matz, C. & Kjelleberg, S. Off the hook—how bacteria survive protozoan grazing. Trends Microbiol. 13, 302–307 (2005).
Hibbing, M. E., Fuqua, C., Parsek, M. R. & Peterson, S. B. Bacterial competition: surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 8, 15–25 (2010).
Dillewijn, P. V., Villadas, P. J. & Toro, N. Effect of a Sinorhizobium meliloti strain with a modified putA gene on the rhizosphere microbial community of alfalfa. Appl. Environ. Microbiol. 68, 4201–4208 (2002).
Iwasaki, K., Uchiyama, H. & Yagi, O. Survival and impact of genetically engineered Pseudomonas putida harboring mercury resistance gene in aquatic microcosms. Biosci. Biotechnol. Biochem. 57, 1264–1269 (1993).
Glandorf, D. C. M. et al. Effect of genetically modified Pseudomonas putida WCS358r on the fungal rhizosphere microflora of field-grown wheat. Appl. Environ. Microbiol. 67, 3371–3378 (2001).
Zhang, X., Nesme, J., Simonet, P. & Frostegård, Å. Fate of invading bacteria in soil and survival of transformants after simulated uptake of transgenes, as evaluated by a model system based on lindane degradation. Res. Microbiol. 163, 200–210 (2012).
Nuti, M. et al. in Ecological Impact of GMO Dissemination in Agro-Ecosystems (eds Lelley, T. et al.) 45–54 (Facultas Verlags-und Buchhandels, 2002).
Ripp, S. et al. Controlled field release of a bioluminescent genetically engineered microorganism for bioremediation process monitoring and control. Environ. Sci. Technol. 34, 846–853 (2000).
Hirsch, P. R. Population dynamics of indigenous and genetically modified rhizobia in the field. N. Phytol. 133, 159–171 (1996).
Corich, V. et al. Long term evaluation of field-released genetically modified rhizobia. Environ. Biosaf. Res. 6, 167–181 (2007).
McBee, R. M. et al. Engineering living and regenerative fungal–bacterial biocomposite structures. Nat. Mater. 21, 471–478 (2022).
De Leij, F. A. A. M., Sutton, E. J., Whipps, J. M., Fenlon, J. S. & Lynch, J. M. Field release of a genetically modified Pseudomonas fluorescens on wheat: establishment, survival and dissemination. Nat. Biotechnol. 13, 1488–1492 (1995).
Wiehe, W. & Höflich, G. Survival of plant growth promoting rhizosphere bacteria in the rhizosphere of different crops and migration to non-inoculated plants under field conditions in north-east Germany. Microbiol. Res. 150, 201–206 (1995).
Kluepfel, D., Kline, E., Skipper, H., Hughes, T. & Gooden, D. The release and tracking of genetically engineered bacteria in the environment. Phytopathology 81, 348–352 (1991).
Jäderlund, L., Hellman, M., Sundh, I., Bailey, M. J. & Jansson, J. K. Use of a novel nonantibiotic triple marker gene cassette to monitor high survival of Pseudomonas fluorescens SBW25 on winter wheat in the field. FEMS Microbiol. Ecol. 63, 156–168 (2008).
Shaw, J. J., Dane, F., Geiger, D. & Kloepper, J. W. Use of bioluminescence for detection of genetically engineered microorganisms released into the environment. Appl. Environ. Microbiol. 58, 267–273 (1992).
Brown, G. G., Doube, B. M. & Edwards, C. Functional interactions between earthworms, microorganisms, organic matter and plants. Earthworm Ecol. 2, 213–239 (2004).
Singer, A. C., Jury, W., Luepromchai, E., Yahng, C. S. & Crowley, D. E. Contribution of earthworms to PCB bioremediation. Soil Biol. Biochem. 33, 765–776 (2001).
Abu-Ashour, J. & Lee, H. Transport of bacteria on sloping soil surfaces by runoff. Environ. Toxicol. 15, 149–153 (2000).
Meola, M., Lazzaro, A. & Zeyer, J. Bacterial composition and survival on Sahara dust particles transported to the European Alps. Front. Microbiol. 6, 1454 (2015).
Gorbushina, A. A. et al. Life in Darwin’s dust: intercontinental transport and survival of microbes in the nineteenth century. Environ. Microbiol. 9, 2911–2922 (2007).
Kolmer, J. A. Tracking wheat rust on a continental scale. Curr. Opin. Plant Biol. 8, 441–449 (2005).
Ford, C. Z., Sayler, G. S. & Burlage, R. S. Containment of a genetically engineered microorganism during a field bioremediation application. Appl. Microbiol. Biotechnol. 51, 397–400 (1999).
Mancinelli, R. L. & Shulls, W. A. Airborne bacteria in an urban environment. Appl. Environ. Microbiol. 35, 1095–1101 (1978).
Tong, X., Leung, M. H., Wilkins, D. & Lee, P. K. City-scale distribution and dispersal routes of mycobiome in residences. Microbiome 5, 131 (2017).
Gattinger, D., Schlenz, V., Weil, T. & Sattler, B. From remote to urbanized: dispersal of antibiotic-resistant bacteria under the aspect of anthropogenic influence. Sci. Total Environ. 924, 171532 (2024).
Mhuireach, G. et al. Urban greenness influences airborne bacterial community composition. Sci. Total Environ. 571, 680–687 (2016).
Zurek, L. & Ghosh, A. Insects represent a link between food animal farms and the urban environment for antibiotic resistance traits. Appl. Environ. Microbiol. 80, 3562–3567 (2014).
Arnold, K. E., Williams, N. J. & Bennett, M. ‘Disperse abroad in the land’: the role of wildlife in the dissemination of antimicrobial resistance. Biol. Lett. 12, 20160137 (2016).
Bankhead, S. B., Landa, B. B., Lutton, E., Weller, D. M. & Gardener, B. B. M. Minimal changes in rhizobacterial population structure following root colonization by wild type and transgenic biocontrol strains. FEMS Microbiol. Ecol. 49, 307–318 (2004).
Lindow, S. E. Competitive exclusion of epiphytic bacteria by Ice−Pseudomonas syringae mutants. Appl. Environ. Microbiol. 53, 2520–2527 (1987).
Schwieger, F. & Tebbe, C. C. Effect of field inoculation with Sinorhizobium meliloti L33 on the composition of bacterial communities in rhizospheres of a target plant Medicago sativa and a non-target plant Chenopodium album—linking of 16S rRNA gene-based single-strand conformation polymorphism community profiles to the diversity of cultivated bacteria. Appl. Environ. Microbiol. 66, 3556–3565 (2000).
Xiong, M., Hu, Z., Zhang, Y., Cheng, X. & Li, C. Survival of GFP-tagged Rhodococcus sp. D310-1 in chlorimuron-ethyl-contaminated soil and its effects on the indigenous microbial community. J. Hazard. Mater. 252-253, 347–354 (2013).
Mawarda, P. C., Le Roux, X., Dirk van Elsas, J. & Salles, J. F. Deliberate introduction of invisible invaders: a critical appraisal of the impact of microbial inoculants on soil microbial communities. Soil Biol. Biochem. 148, 107874 (2020).
Philippot, L., Griffiths, B. S. & Langenheder, S. Microbial community resilience across ecosystems and multiple disturbances. Microbiol. Mol. Biol. Rev. https://doi.org/10.1128/mmbr.00026-20 (2021).
Shade, A. et al. Fundamentals of microbial community resistance and resilience. Front. Microbiol. 3, 417 (2012).
Lawson, C. E. et al. Common principles and best practices for engineering microbiomes. Nat. Rev. Microbiol. 17, 725–741 (2019).
Gómez Expósito, R., de Bruijn, I., Postma, J. & Raaijmakers, J. M. Current insights into the role of rhizosphere bacteria in disease suppressive soils. Front. Microbiol. 8, 2529 (2017).
van Elsas, J. D. et al. Microbial diversity determines the invasion of soil by a bacterial pathogen. Proc. Natl Acad. Sci. USA 109, 1159–1164 (2012).
Gregory, A. C. et al. Marine DNA viral macro- and microdiversity from pole to pole. Cell 177, 1109–1123.e1114 (2019).
Rizzo, L. et al. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: a review. Sci. Total Environ. 447, 345–360 (2013).
Andersen, J. T., Schäfer, T., Jørgensen, P. L. & Møller, S. Using inactivated microbial biomass as fertilizer: the fate of antibiotic resistance genes in the environment. Res. Microbiol. 152, 823–833 (2001).
Peters, M. et al. Acquisition of a deliberately introduced phenol degradation operon, pheBA, by different indigenous Pseudomonas species. Appl. Environ. Microbiol. 63, 4899–4906 (1997).
National Research Council; Division on Earth and Life Studies; Commission on Life Sciences; Committee on Scientific Evaluation of the Introduction of Genetically Modified Microorganisms and Plants into the Environment. Field Testing Genetically Modified Organisms: Framework for Decisions (The National Academies Press, 1989).
Lilley, A. K. & Bailey, M. J. The acquisition of indigenous plasmids by a genetically marked pseudomonad population colonizing the sugar beet phytosphere is related to local environmental conditions. Appl. Environ. Microbiol. 63, 1577–1583 (1997).
Munck, C., Sheth, R. U., Freedberg, D. E. & Wang, H. H. Recording mobile DNA in the gut microbiota using an Escherichia coli CRISPR-Cas spacer acquisition platform. Nat. Commun. 11, 95 (2020).
Allgeier, S., Friedrich, A. & Brühl, C. A. Mosquito control based on Bacillus thuringiensis israelensis (Bti) interrupts artificial wetland food chains. Sci. Total Environ. 686, 1173–1184 (2019).
Hernández-Rodríguez, C. S., de Escudero, I. R., Asensio, A. C., Ferré, J. & Caballero, P. Encapsulation of the Bacillus thuringiensis secretable toxins Vip3Aa and Cry1Ia in Pseudomonas fluorescens. Biol. Control 66, 159–165 (2013).
Code of Federal Regulations, CFR § 158.2150—Microbial Pesticides Nontarget Organisms and Environmental Fate Data Requirements Table (National Archives, 2015); https://www.ecfr.gov/current/title-40/chapter-I/subchapter-E/part-158/subpart-V/section-158.2150 (2015).
US Environmental Protection Agency. OCSPP Harmonized Test Guidelines—Master List (US EPA, 2019); https://www.epa.gov/sites/default/files/2019-10/documents/ocspp-testguidelines_masterlist-2019-09-24.pdf
Rathinam, M., Singh, S., Pattanayak, D. & Sreevathsa, R. Comprehensive in silico allergenicity assessment of novel protein engineered chimeric Cry proteins for safe deployment in crops. BMC Biotechnol. 17, 64 (2017).
Wozniak, C. & Kough, J. Regulation of transgenic forage grasses under the proposed plant-pesticide rule. In Proc. Thirty Sixth Grass Breeders’ Work Planning Conference 18–24 (The Noble Foundation, 2000).
Environmental Protection Agency. Status Report: TSCA Biotechnology Submissions for Fiscal Years 87–98 (US EPA, 1999); https://www.epa.gov/sites/default/files/2016-05/documents/bistat98.pdf
Miller, T. A. in Regulation of Agricultural Biotechnology: The United States and Canada (eds Wozniak, C. A. & McHughen, A.) 103–122 (Springer, 2012).
Kerr, A. & Bullard, G. Biocontrol of crown gall by Rhizobium rhizogenes: challenges in biopesticide commercialisation. Agronomy 10, 1126 (2020).
Arana, I. et al. Effect of temperature and starvation upon survival strategies of Pseudomonas fluorescens CHA0: comparison with Escherichia coli. FEMS Microbiol. Ecol. 74, 500–509 (2010).
Winnenburg, R. et al. PHI-base: a new database for pathogen host interactions. Nucleic Acids Res. 34, D459–D464 (2006).
Bartoszewicz, J. M., Seidel, A., Rentzsch, R. & Renard, B. Y. DeePaC: predicting pathogenic potential of novel DNA with reverse-complement neural networks. Bioinformatics 36, 81–89 (2020).
Cosentino, S., Voldby Larsen, M., Møller Aarestrup, F. & Lund, O. PathogenFinder—distinguishing friend from foe using bacterial whole genome sequence data. PLoS ONE 8, e77302 (2013).
Sperschneider, J. Machine learning in plant-pathogen interactions: empowering biological predictions from field scale to genome scale. N. Phytol. 228, 35–41 (2020).
Hays, S. G., Patrick, W. G., Ziesack, M., Oxman, N. & Silver, P. A. Better together: engineering and application of microbial symbioses. Curr. Opin. Biotechnol. 36, 40–49 (2015).
McCarty, N. S. & Ledesma-Amaro, R. Synthetic biology tools to engineer microbial communities for biotechnology. Trends Biotechnol. 37, 181–197 (2019).
Pivot Bio. PROVEN40 Safety Data Sheet (Pivot Bio, 2023); https://www.pivotbio.com/hubfs/Safety%20Data%20Sheets/2022%20SDS-Pivot%20Bio%20PROVEN40%20LIF.pdf
Baghapour, M. A., Nasseri, S. & Derakhshan, Z. Atrazine removal from aqueous solutions using submerged biological aerated filter. J. Environ. Health Sci. Eng. 11, 6 (2013).
Mnif, I. et al. Biodegradation of diesel oil by a novel microbial consortium: comparison between co-inoculation with biosurfactant-producing strain and exogenously added biosurfactants. Environ. Sci. Pollut. Res. 22, 14852–14861 (2015).
Rawlings, D. E. & Johnson, D. B. The microbiology of biomining: development and optimization of mineral-oxidizing microbial consortia. Microbiology 153, 315–324 (2007).
Cheng, A. G. et al. Design, construction and in vivo augmentation of a complex gut microbiome. Cell https://doi.org/10.1016/j.cell.2022.08.003 (2022).
Bhagwat, A. A. & Keister, D. L. Improved inoculant strains of Bradyrhizobium japonicum. PCT patent WO2001038492A1 (2000).
Halperin, S. O. et al. CRISPR-guided DNA polymerases enable diversification of all nucleotides in a tunable window. Nature 560, 248–252 (2018).
Zhang, Y.-X. et al. Genome shuffling leads to rapid phenotypic improvement in bacteria. Nature 415, 644–646 (2002).
Gong, J., Zheng, H., Wu, Z., Chen, T. & Zhao, X. Genome shuffling: progress and applications for phenotype improvement. Biotechnol. Adv. 27, 996–1005 (2009).
Bloch, S. E. et al. Biological nitrogen fixation in maize: optimizing nitrogenase expression in a root-associated diazotroph. J. Exp. Bot. 71, 4591–4603 (2020).
Jones, D. A., Ryder, M. H., Clare, B. G., Farrand, S. K. & Kerr, A. Construction of a Tra− deletion mutant of pAgK84 to safeguard the biological control of crown gall. Mol. Gen. Genet. MGG 212, 207–214 (1988).
Frost, L. S., Leplae, R., Summers, A. O. & Toussaint, A. Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 3, 722–732 (2005).
US Environmental Protection Agency. TSCA Biotechnology Notifications Status for Cases Reviewed Prior to June 22, 2016 (US EPA, 2024); https://www.epa.gov/regulation-biotechnology-under-tsca-and-fifra/tsca-biotechnology-notifications-status-cases
Wen, A. et al. Enabling biological nitrogen fixation for cereal crops in fertilized fields. ACS Synth. Biol. 10, 3264–3277 (2021).
Hmelo, L. R. et al. Precision-engineering the Pseudomonas aeruginosa genome with two-step allelic exchange. Nat. Protoc. 10, 1820–1841 (2015).
Scott, M., Gunderson, C. W., Mateescu, E. M., Zhang, Z. & Hwa, T. Interdependence of cell growth and gene expression: origins and consequences. Science 330, 1099–1102 (2010).
Sleight, S. C., Bartley, B. A., Lieviant, J. A. & Sauro, H. M. Designing and engineering evolutionary robust genetic circuits. J. Biol. Eng. 4, 12 (2010).
Shen, P. & Huang, H. V. Homologous recombination in Escherichia coli: dependence on substrate length and homology. Genetics 112, 441–457 (1986).
Menuhin-Gruman, I. et al. Evolutionary Stability Optimizer (ESO): a novel approach to identify and avoid mutational hotspots in DNA sequences while maintaining high expression levels. ACS Synth. Biol. 11, 1142–1151 (2022).
Jack, B. R. et al. Predicting the genetic stability of engineered DNA sequences with the EFM calculator. ACS Synth. Biol. 4, 939–943 (2015).
Blazejewski, T., Ho, H.-I. & Wang, H. H. Synthetic sequence entanglement augments stability and containment of genetic information in cells. Science 365, 595–598 (2019).
Posfai, G. et al. Emergent properties of reduced-genome Escherichia coli. Science 312, 1044–1046 (2006).
Csörgő, B., Fehér, T., Tímár, E., Blattner, F. R. & Pósfai, G. Low-mutation-rate, reduced-genome Escherichia coli: an improved host for faithful maintenance of engineered genetic constructs. Microb. Cell Factories 11, 11 (2012).
Unthan, S. et al. Chassis organism from Corynebacterium glutamicum–a top‐down approach to identify and delete irrelevant gene clusters. Biotechnol. J. 10, 290–301 (2015).
Komatsu, M., Uchiyama, T., Ōmura, S., Cane, D. E. & Ikeda, H. Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism. Proc. Natl Acad. Sci. USA 107, 2646–2651 (2010).
Shen, X. et al. Developing genome-reduced Pseudomonas chlororaphis strains for the production of secondary metabolites. BMC Genomics 18, 715 (2017).
Reuß, D. R., Commichau, F. M., Gundlach, J., Zhu, B. & Stülke, J. The blueprint of a minimal cell: MiniBacillus. Microbiol. Mol. Biol. Rev. 80, 955–987 (2016).
Sturino, J. M. & Klaenhammer, T. R. Engineered bacteriophage-defence systems in bioprocessing. Nat. Rev. Microbiol. 4, 395–404 (2006).
Suttle, C. A. Marine viruses–major players in the global ecosystem. Nat. Rev. Microbiol. 5, 801–812 (2007).
Ashelford, K. E., Day, M. J. & Fry, J. C. Elevated abundance of bacteriophage infecting bacteria in soil. Appl. Environ. Microbiol. 69, 285–289 (2003).
Ofir, G. et al. DISARM is a widespread bacterial defence system with broad anti-phage activities. Nat. Microbiol. 3, 90–98 (2018).
Lopatina, A., Tal, N. & Sorek, R. Abortive infection: bacterial suicide as an antiviral immune strategy. Annu. Rev. Virol. 7, 371–384 (2020).
Shen, Q., Zhou, X.-T., Guo, Q., Xue, Y.-P. & Zheng, Y.-G. Engineering laboratory/factory-specific phage-resistant strains of Escherichia coli by mutagenesis and screening. World J. Microbiol. Biotechnol. 38, 51 (2022).
Mutalik, V. K. et al. High-throughput mapping of the phage resistance landscape in E. coli. PLoS Biol. 18, e3000877 (2020).
Tan, D. et al. A frameshift mutation in wcaJ associated with phage resistance in Klebsiella pneumoniae. Microorganisms 8, 378 (2020).
Cai, R. et al. A smooth-type, phage-resistant Klebsiella pneumoniae mutant strain reveals that OmpC is indispensable for infection by phage GH-K3. Appl. Environ. Microbiol. 84, e01585–01518 (2018).
Zheng, K. et al. Highly efficient base editing in bacteria using a Cas9-cytidine deaminase fusion. Commun. Biol. 1, 32 (2018).
Hutton, P. O., Schneider, W. R. & Rispin, A. S. US EPA, Office of Pesticide Programs Memorandum: Classification of Killed Microbial Pesticides (US EPA, 1989).
Strong, L. C., McTavish, H., Sadowsky, M. J. & Wackett, L. P. Field-scale remediation of atrazine-contaminated soil using recombinant Escherichia coli expressing atrazine chlorohydrolase. Environ. Microbiol. 2, 91–98 (2000).
Fan, C. et al. Chromosome-free bacterial cells are safe and programmable platforms for synthetic biology. Proc. Natl Acad. Sci. USA 117, 6752–6761 (2020).
Qian, J. et al. Barcoded microbial system for high-resolution object provenance. Science 368, 1135–1140 (2020).
Silverman, A. D., Akova, U., Alam, K. K., Jewett, M. C. & Lucks, J. B. Design and optimization of a cell-free atrazine biosensor. ACS Synth. Biol. 9, 671–677 (2020).
Chemla, Y., Ozer, E., Schlesinger, O., Noireaux, V. & Alfonta, L. Genetically expanded cell-free protein synthesis using endogenous pyrrolysyl orthogonal translation system. Biotechnol. Bioeng. 112, 1663–1672 (2015).
Pardee, K. et al. Rapid, low-cost detection of Zika virus using programmable biomolecular components. Cell 165, 1255–1266 (2016).
Tang, T.-C. et al. Hydrogel-based biocontainment of bacteria for continuous sensing and computation. Nat. Chem. Biol. 17, 724–731 (2021).
Lin, G.-M. & Voigt, C. A. Design of a redox-proficient Escherichia coli for screening terpenoids and modifying cytochrome P450s. Nat. Catal. 6, 1016–1029 (2023).
Sayler, G. S. et al. Field Application of a Genetically Engineered Microorganism for Polycyclic Aromatic Hydrocarbon Bioremediation Process Monitoring and Control (Springer, 1999).
Belkin, S. et al. Remote detection of buried landmines using a bacterial sensor. Nat. Biotechnol. 35, 308–310 (2017).
Gallagher, R. R., Patel, J. R., Interiano, A. L., Rovner, A. J. & Isaacs, F. J. Multilayered genetic safeguards limit growth of microorganisms to defined environments. Nucleic Acids Res. 43, 1945–1954 (2015).
Steidler, L. et al. Biological containment of genetically modified Lactococcus lactis for intestinal delivery of human interleukin 10. Nat. Biotechnol. 21, 785–789 (2003).
Adolfsen, K. J. et al. Improvement of a synthetic live bacterial therapeutic for phenylketonuria with biosensor-enabled enzyme engineering. Nat. Commun. 12, 6215 (2021).
Lubkowicz, D. et al. An engineered bacterial therapeutic lowers urinary oxalate in preclinical models and in silico simulations of enteric hyperoxaluria. Mol. Syst. Biol. 18, e10539 (2022).
Arnaouteli, S., Bamford, N. C., Stanley-Wall, N. R. & Kovács, Á. T. Bacillus subtilis biofilm formation and social interactions. Nat. Rev. Microbiol. 19, 600–614 (2021).
US Environmental Protection Agency Office of Inspector General. Science to Support Rulemaking Report 2003-P-00003 (addendum) (US EPA, 2002); https://www.epa.gov/sites/default/files/2015-12/documents/ssrulemakingaddendum.pdf
US Environmental Protection Agency Office of Pollution Prevention and Toxics. TSCA Experimental Release Application Approved for Pseudomonas putida Strains fact sheet (US EPA); https://www.epa.gov/regulation-biotechnology-under-tsca-and-fifra/tsca-experimental-release-application-approved-0
Sikorski, J., Graupner, S., Lorenz, M. G. & Wackernagel, W. Natural genetic transformation of Pseudomonas stutzeri in a non-sterile soil. Microbiology 144, 569–576 (1998).
Asin-Garcia, E. et al. Phosphite synthetic auxotrophy as an effective biocontainment strategy for the industrial chassis Pseudomonas putida. Microb. Cell Fact. 21, 156 (2022).
Gay, P., Le Coq, D., Steinmetz, M., Berkelman, T. & Kado, C. Positive selection procedure for entrapment of insertion sequence elements in gram-negative bacteria. J. Bacteriol. 164, 918–921 (1985).
Hayashi, N., Lai, Y., Fuerte-Stone, J., Mimee, M. & Lu, T. K. Cas9-assisted biological containment of a genetically engineered human commensal bacterium and genetic elements. Nat. Commun. 15, 2096 (2024).
Lee, J. W., Chan, C. T. Y., Slomovic, S. & Collins, J. J. Next-generation biocontainment systems for engineered organisms. Nat. Chem. Biol. 14, 530–537 (2018).
Kim, D. & Lee, J. W. Genetic biocontainment systems for the safe use of engineered microorganisms. Biotechnol. Bioprocess Eng. 25, 974–984 (2020).
Ishikawa, M., Kojima, T. & Hori, K. Development of a biocontained toluene-degrading bacterium for environmental protection. Microbiol. Spectrum 9, 10.1128/spectrum.00259-00221 (2021).
Molin, S. et al. Conditional suicide system for containment of bacteria and plasmids. Nat. Biotechnol. 5, 1315–1318 (1987).
Contreras, A., Molin, S. & Ramos, J.-L. Conditional-suicide containment system for bacteria which mineralize aromatics. Appl. Environ. Microbiol. 57, 1504–1508 (1991).
Bej, A. K., Perlin, M. H. & Atlas, R. M. Model suicide vector for containment of genetically engineered microorganisms. Appl. Environ. Microbiol. 54, 2472–2477 (1988).
Rottinghaus, A. G., Ferreiro, A., Fishbein, S. R. S., Dantas, G. & Moon, T. S. Genetically stable CRISPR-based kill switches for engineered microbes. Nat. Commun. 13, 672 (2022).
Caliando, B. J. & Voigt, C. A. Targeted DNA degradation using a CRISPR device stably carried in the host genome. Nat. Commun. 6, 6989 (2015).
Chan, C. T. Y., Lee, J. W., Cameron, D. E., Bashor, C. J. & Collins, J. J. ‘Deadman’ and ‘Passcode’ microbial kill switches for bacterial containment. Nat. Chem. Biol. 12, 82–86 (2016).
Halvorsen, T. M., Ricci, D. P., Park, D. M., Jiao, Y. & Yung, M. C. Comparison of kill switch toxins in plant-beneficial Pseudomonas fluorescens reveals drivers of lethality, stability and escape. ACS Synth. Biol. 11, 3785–3796 (2022).
Yaung, S. J. et al. Improving microbial fitness in the mammalian gut by in vivo temporal functional metagenomics. Mol. Syst. Biol. 11, 788 (2015).
Kong, W. et al. Regulated programmed lysis of recombinant Salmonella in host tissues to release protective antigens and confer biological containment. Proc. Natl Acad. Sci. USA 105, 9361–9366 (2008).
Garson, J. et al. Detection of hepatitis C viral sequences in blood donations by ‘nested’ polymerase chain reaction and prediction of infectivity. Lancet 335, 1419–1422 (1990).
Jasinska, W. et al. Chromosomal barcoding of E. coli populations reveals lineage diversity dynamics at high resolution. Nat. Ecol. Evol. 4, 437–452 (2020).
Ahrenholtz, I., Lorenz, M. G. & Wackernagel, W. A conditional suicide system in Escherichia coli based on the intracellular degradation of DNA. Appl. Environ. Microbiol. 60, 3746–3751 (1994).
Zürcher, J. F. et al. Refactored genetic codes enable bidirectional genetic isolation. Science 378, 516–523 (2022).
Liu, C. C., Jewett, M. C., Chin, J. W. & Voigt, C. A. Toward an orthogonal central dogma. Nat. Chem. Biol. 14, 103–106 (2018).
Rackham, O. & Chin, J. W. A network of orthogonal ribosome·mRNA pairs. Nat. Chem. Biol. 1, 159–166 (2005).
Schmidt, M. & de Lorenzo, V. Synthetic constructs in/for the environment: managing the interplay between natural and engineered Biology. FEBS Lett. 586, 2199–2206 (2012).
Zhang, Y. et al. A semi-synthetic organism that stores and retrieves increased genetic information. Nature 551, 644–647 (2017).
Fan, C., Deng, Q. & Zhu, T. F. Bioorthogonal information storage in l-DNA with a high-fidelity mirror-image Pfu DNA polymerase. Nat. Biotechnol. https://doi.org/10.1038/s41587-021-00969-6 (2021).
Xu, Y. & Zhu, T. F. Mirror-image T7 transcription of chirally inverted ribosomal and functional RNAs. Science 378, 405–412 (2022).
Schmidt, M. & de Lorenzo, V. Synthetic bugs on the loose: containment options for deeply engineered (micro) organisms. Curr. Opin. Biotechnol. 38, 90–96 (2016).
Sanders, M. E. & Klaenhammer, T. R. Characterization of phage-sensitive mutants from a phage-insensitive strain of Streptococcus lactis: evidence for a plasmid determinant that prevents phage adsorption. Appl. Environ. Microbiol. 46, 1125–1133 (1983).
Zou, X. et al. Systematic strategies for developing phage resistant Escherichia coli strains. Nat. Commun. 13, 4491 (2022).
Vassallo, C. N., Doering, C. R., Littlehale, M. L., Teodoro, G. I. C. & Laub, M. T. A functional selection reveals previously undetected anti-phage defence systems in the E. coli pangenome. Nat. Microbiol. https://doi.org/10.1038/s41564-022-01219-4 (2022).
Fredens, J. et al. Total synthesis of Escherichia coli with a recoded genome. Nature 569, 514–518 (2019).
Lajoie, M. J. et al. Genomically recoded organisms expand biological functions. Science 342, 357–360 (2013).
Robertson, W. E. et al. Sense codon reassignment enables viral resistance and encoded polymer synthesis. Science 372, 1057–1062 (2021).
Nyerges, A. et al. A swapped genetic code prevents viral infections and gene transfer. Nature 615, 720–727 (2023).
Johns, N. I., Blazejewski, T., Gomes, A. L. C. & Wang, H. H. Principles for designing synthetic microbial communities. Curr. Opin. Microbiol. 31, 146–153 (2016).
Kylilis, N., Tuza, Z. A., Stan, G.-B. & Polizzi, K. M. Tools for engineering coordinated system behaviour in synthetic microbial consortia. Nat. Commun. 9, 2677 (2018).
Gupta, S. et al. Investigating the dynamics of microbial consortia in spatially structured environments. Nat. Commun. 11, 2418 (2020).
Qian, Y., Lan, F. & Venturelli, O. S. Towards a deeper understanding of microbial communities: integrating experimental data with dynamic models. Curr. Opin. Microbiol. 62, 84–92 (2021).
Ren, X. & Murray, R. M. Cooperation enhances robustness of coexistence in spatially structured consortia. In Proc. 2019 18th European Control Conference (ECC) (eds Garofalo, F. et al.) 2651–2656 (IEEE, 2019).
Kiers, E. T., Hutton, M. G. & Denison, R. F. Human selection and the relaxation of legume defences against ineffective rhizobia. Proc. R. Soc. B Biol. Sci. 274, 3119–3126 (2007).
Bulgarelli, D., Schlaeppi, K., Spaepen, S., Themaat, E. V. L. V. & Schulze-Lefert, P. Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 64, 807–838 (2013).
Haskett, T. L. et al. Engineered plant control of associative nitrogen fixation. Proc. Natl Acad. Sci. USA 119, e2117465119 (2022).
Sayler, G. S. & Ripp, S. Field applications of genetically engineered microorganisms for bioremediation processes. Curr. Opin. Biotechnol. 11, 286–289 (2000).
Wilson, M. & Lindow, S. Release of recombinant microorganisms. Annu. Rev. Microbiol. 47, 913–944 (1993).
Sheahan, T. & Wieden, H.-J. Emerging regulatory challenges of next-generation synthetic biology. Biochem. Cell Biol. 99, 766–771 (2021).
US Office of Science and Technology Policy. Coordinated framework for regulation of biotechnology. Fed. Regist. 51, 23302-50 (1986).
Marken, J. P., Maxon, M. E. & Murray, R. M. Policy Recommendations for the Regulation of Engineered Microbes for Environmental Release (Linde Center for Science, Society and Policy, Caltech, 2024); https://doi.org/10.57959/bgny-v542
Marchant, G. E. The Growing Gap between Emerging Technologies and the Law (Springer, 2011).
French, K. E., Zhou, Z. & Terry, N. Horizontal ‘gene drives’ harness indigenous bacteria for bioremediation. Sci. Rep. 10, 15091 (2020).
Mutalik, V. K. et al. Precise and reliable gene expression via standard transcription and translation initiation elements. Nat. Methods 10, 354–360 (2013).
Reichman, J. R. et al. Research Needs for Novel Engineered Microbes & Biopesticides Intended for Open Release into the Environment. Report EPA/600/R-22/163 (US EPA, 2022).
Warner, C. M. et al. in Synthetic Biology 2020: Frontiers in Risk Analysis and Governance (eds Trump, B. D. et al.) 19–50 (Springer, 2020).
US Department of Agriculture—Animal and Plant Health Inspection Service. APHIS eFile: New SOP Template for Modified Microbe Movement Permits (US Department of Agriculture, 2024); https://www.aphis.usda.gov/news/program-update/aphis-efile-new-sop-template-modified-microbe-movement-permits
US Department of Agriculture—Animal and Plant Health Inspection Service. Draft: Guide for Submitting Permit Applications for Microorganisms Developed Using Genetic Engineering Under 7 CFR Part 340 (US Department of Agriculture, 2023); https://www.aphis.usda.gov/biotechnology/downloads/draft-brs-microbe-permit-guide.pdf
US Congress National Security Commission on Emerging Biotechnology. Interim Report—December 2023 (National Security Commission on Emerging Biotechnology, 2023); https://www.biotech.senate.gov/wp-content/uploads/2024/01/Biotech-Commission-Dec2023-Report.pdf
Defense Advanced Reseach Projects Agency. A Scent-Based Strategy for Preventing Mosquito Transmission of Disease (DARPA, 2019); https://www.darpa.mil/news-events/2019-05-03a#:~:text=ReVector%2C%20a%20new%20program%20from,to%20temporarily%20alter%20chemical%20production
Rosewitz, J. A., Wang, S., Scarlata, S. F. & Rahbar, N. An enzymatic self-healing cementitious material. Appl. Mater. Today 23, 101035 (2021).
Xu, Y. & Lu, M. Microbially enhanced oil recovery at simulated reservoir conditions by use of engineered bacteria. J. Pet. Sci. Eng. 78, 233–238 (2011).
Chen, Y. E. et al. Engineered skin bacteria induce antitumor T cell responses against melanoma. Science 380, 203–210 (2023).
Becker, M. H. et al. Towards a better understanding of the use of probiotics for preventing chytridiomycosis in Panamanian golden frogs. Ecohealth 8, 501–506 (2011).
Forkus, B., Ritter, S., Vlysidis, M., Geldart, K. & Kaznessis, Y. N. Antimicrobial probiotics reduce Salmonella enterica in Turkey gastrointestinal tracts. Sci. Rep. 7, 40695 (2017).
US Department of Agriculture—Animal and Plant Health Inspection Service. Importation, Interstate Movement and Environmental Release of Certain Genetically Engineered Organisms (US Department of Agriculture, 2017); https://www.govinfo.gov/content/pkg/FR-2017-01-19/pdf/2017-00858.pdf
US Food & Drug Administration. FDA Organization Charts (US FDA, 2019); https://www.fda.gov/about-fda/fda-organization/fda-organization-charts
Environmental Protection Agency—Office of Pollution Prevention and Toxics. Overview of Biotechnology Under TSCA (US EPA, 2023); https://www.epa.gov/regulation-biotechnology-under-tsca-and-fifra/overview-biotechnology-under-tsca
Lindow, S. E. & Panopoulos, N. Field tests of recombinant Ice− Pseudomonas syringae for biological frost control in potato. In Proc. First International Conference on the Release of Genetically-engineered Micro-organisms (eds Sussman, M. et al.) 121–138 (Academic, 1988).
Dillewijn, P. V., Soto, M. A. J., Villadas, P. J. & Toro, N. Construction and environmental release of a Sinorhizobium meliloti strain genetically modified to be more competitive for alfalfa nodulation. Appl. Environ. Microbiol. 67, 3860–3865 (2001).
Viebahn, M. et al. Repeated introduction of genetically modified Pseudomonas putida WCS358r without intensified effects on the indigenous microflora of field-grown wheat. Appl. Environ. Microbiol. 69, 3110–3118 (2003).
O’flaherty, S. et al. Greenhouse and field evaluations of an autoselective system based on an essential thymidylate synthase gene for improved maintenance of plasmid vectors in modified Rhizobium meliloti. Appl. Environ. Microbiol. 61, 4051–4056 (1995).
Vicedo, B., Peñalver, R., Asins, M. J. & López, M. M. Biological control of Agrobacterium tumefaciens, colonization and pAgK84 transfer with Agrobacterium radiobacter K84 and the Tra− mutant strain K1026. Appl. Environ. Microbiol. 59, 309–315 (1993).
Parakhonsky, E. ОСОБЕННОСТИ РАЗВИТИЯ, ТУШЕНИЯ И ЛИКВИДАЦИИ ПОСЛЕДСТВИЙ ПОДЗЕМНЫХ ПОЖАРОВ НА СЛАНЦЕВЫХ ШАХТАХ. Oil Shale 12, 63–77 (1995).
Das, J. & Dangar, T. in Microbial Biotechnology: Methods and Applications (eds Mishra, B. B. & Thatoi, H.) 68–95 (Narosa Publishing House, 2011).
Charnley, A. Microbial pathogens and insect pest control. Lett. Appl. Microbiol. 12, 149–157 (1991).
Menegat, S., Ledo, A. & Tirado, R. Greenhouse gas emissions from global production and use of nitrogen synthetic fertilisers in agriculture. Sci. Rep. 12, 14490 (2022).
Pivot Bio. 2022 Impact Report (Pivot Bio, 2023); https://www.pivotbio.com/hubfs/1604%20-%20US%20Website%20-%20Impact%20Report%20LP/Pivot%20Bio%202022%20Impact%20Report.pdf
Hillman, J. D. Genetically modified Streptococcus mutans for the prevention of dental caries. Antonie Van Leeuwenhoek 82, 361–366 (2002).
Acknowledgements
We thank L. Canfora, B. Adams, N. Hubbard and I. Varkovitzsky for their insightful discussions. C. Voigt is supported as a Schmidt Futures Fellow. The work was also supported by the MIT Climate Grand Challenge program, the Ministry of Defense of Israel (MIT 4441024394) and by the Army Research Office and was accomplished under Cooperative Agreement no. W911NF-22-2-0210 (CHARMME). The views and conclusions contained in this document are those of the authors and should not be interpreted as representing the official policies, either expressed or implied, of the Army Research Office or the US Government. The US Government is authorized to reproduce and distribute reprints for Government purposes notwithstanding any copyright notation herein.
Author information
Authors and Affiliations
Contributions
Y.C., C.J.S., C.A.W. and C.A.V. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
Y.C. declares no competing interests. C.A.V. is a founder of Pivot Bio. C.J.S. is a founder of Robigo, Inc., and C.A.V. serves on their Scientific Advisory Board. C.A.V. is a founder and Scientific Advisory Board member of Fieldstone Bio. C.A.W. used to work at the US Environmental Protection Agency but is now retired.
Peer review
Peer review information
Nature Microbiology thanks Matthew Sorbara, Yasuo Yoshikuni and Robert Egbert for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary notes 1–4 and Tables 1–5.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chemla, Y., Sweeney, C.J., Wozniak, C.A. et al. Design and regulation of engineered bacteria for environmental release. Nat Microbiol 10, 281–300 (2025). https://doi.org/10.1038/s41564-024-01918-0
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41564-024-01918-0
This article is cited by
-
Harnessing bacterial consortia for effective bioremediation: targeted removal of heavy metals, hydrocarbons, and persistent pollutants
Environmental Sciences Europe (2025)
-
Hyperspectral reporters for long-distance and wide-area detection of gene expression in living bacteria
Nature Biotechnology (2025)
-
Standardization guidelines and future trends for PET hydrolase research
Nature Communications (2025)
-
Occurrence and microbial remediation of polycyclic aromatic hydrocarbons and heavy metals pollution in soils
World Journal of Microbiology and Biotechnology (2025)
-
Engineering biology applications for environmental solutions: potential and challenges
Nature Communications (2025)