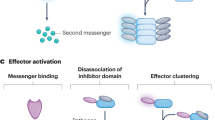
Abstract
The use of whole-genome sequencing to monitor bacterial pathogens has provided crucial insights into their within-host evolution, revealing mutagenic and selective processes driving the emergence of antibiotic resistance, immune evasion phenotypes and adaptations that enable sustained human-to-human transmission. Deep genomic and metagenomic sequencing of intra-host pathogen populations is also enhancing our ability to track bacterial transmission, a key component of infection control. This Review discusses the major processes driving bacterial evolution within humans, including both pathogenic and commensal species. Initially, mutational processes, including how mutational signatures reveal pathogen biology, and the selective pressures driving evolution are considered. The dynamics of horizontal gene transfer and intra-host pathogen competition are also examined, followed by a focus on the emergence of bacterial pathogenesis. Finally, the Review focuses on the importance of within-host genetic diversity in tracking bacterial transmission and its implications for infectious disease control and public health.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Bryant, J. M. et al. Stepwise pathogenic evolution of Mycobacterium abscessus. Science 372, eabb8699 (2021).
Lieberman, T. D. et al. Parallel bacterial evolution within multiple patients identifies candidate pathogenicity genes. Nat. Genet. 43, 1275–1280 (2011).
Ruis, C. et al. Mutational spectra are associated with bacterial niche. Nat. Commun. 14, 7091 (2023).
Key, F. M. et al. On-person adaptive evolution of Staphylococcus aureus during treatment for atopic dermatitis. Cell Host Microbe 31, 593–603 (2023).
Duchêne, S. et al. Genome-scale rates of evolutionary change in bacteria. Microb. Genom. 2, e000094 (2016).
Moura de Sousa, J., Lourenço, M. & Gordo, I. Horizontal gene transfer among host-associated microbes. Cell Host Microbe 31, 513–527 (2023).
Valles-Colomer, M. et al. The person-to-person transmission landscape of the gut and oral microbiomes. Nature 614, 125–135 (2023).
Wozniak, K. J. & Simmons, L. A. Bacterial DNA excision repair pathways. Nat. Rev. Microbiol. 20, 465–477 (2022).
Sender, R., Fuchs, S. & Milo, R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 14, e1002533 (2016).
Jorth, P. et al. Regional isolation drives bacterial diversification within cystic fibrosis lungs. Cell Host Microbe 18, 307–319 (2015).
Mahrt, N. et al. Bottleneck size and selection level reproducibly impact evolution of antibiotic resistance. Nat. Ecol. Evol. https://doi.org/10.1038/s41559-021-01511-2 (2021).
Didelot, X. et al. Genomic evolution and transmission of Helicobacter pylori in two South African families. Proc. Natl Acad. Sci. USA 110, 13880–13885 (2013).
Rocha, E. P. C. et al. Comparisons of dN/dS are time dependent for closely related bacterial genomes. J. Theor. Biol. 239, 226–235 (2006).
Smith, E. E. et al. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc. Natl Acad. Sci. USA 103, 8487–8492 (2006).
Weimann, A. et al. Evolution and host-specific adaptation of Pseudomonas aeruginosa. Science 385, eadi0908 (2024).
Marvig, R. L., Sommer, L. M., Molin, S. & Johansen, H. K. Convergent evolution and adaptation of Pseudomonas aeruginosa within patients with cystic fibrosis. Nat. Genet. 47, 57–64 (2015).
Lieberman, T. D. et al. Genomic diversity in autopsy samples reveals within-host dissemination of HIV-associated Mycobacterium tuberculosis. Nat. Med. 22, 1470–1474 (2016).
Zhao, S. et al. Adaptive evolution within gut microbiomes of healthy people. Cell Host Microbe https://doi.org/10.1016/j.chom.2019.03.007 (2019).
Ailloud, F. et al. Within-host evolution of Helicobacter pylori shaped by niche-specific adaptation, intragastric migrations and selective sweeps. Nat. Commun. 10, 2273 (2019).
Kennemann, L. et al. Helicobacter pylori genome evolution during human infection. Proc. Natl Acad. Sci. USA 108, 5033–5038 (2011).
Sassone-Corsi, M. et al. Microcins mediate competition among Enterobacteriaceae in the inflamed gut. Nature 540, 280–283 (2016).
Forsyth, J. H. et al. Decolonizing drug-resistant E. coli with phage and probiotics: breaking the frequency-dependent dominance of residents. Microbiology 169, 001352 (2023).
Ruis, C. et al. A lung-specific mutational signature enables inference of viral and bacterial respiratory niche. Micro. Genom. 9, mgen001018 (2023).
Ruis, C. et al. Dissemination of Mycobacterium abscessus via global transmission networks. Nat. Microbiol. 6, 1279–1288 (2021).
Alexandrov, L. B. et al. Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013).
Alexandrov, L. B. et al. Mutational signatures associated with tobacco smoking in human cancer. Science 354, 618–622 (2016).
Kucab, J. E. et al. A compendium of mutational signatures of environmental agents. Cell 177, 821–836 (2019).
Endutkin, A. V. & Zharkov, D. O. GO system, a DNA repair pathway to cope with oxidative damage. Mol. Biol. 55, 193–210 (2021).
Tonkin-Hill, G. et al. Pneumococcal within-host diversity during colonization, transmission and treatment. Nat. Microbiol. 7, 1791–1804 (2022).
Pleguezuelos-Manzano, C. et al. Mutational signature in colorectal cancer caused by genotoxic pks+ E. coli. Nature 580, 269–273 (2020).
Couce, A., Guelfo, J. R. & Blázquez, J. Mutational spectrum drives the rise of mutator bacteria. PLoS Genet. 9, e1003167 (2013).
Lee, H., Popodi, E., Tang, H. & Foster, P. L. Rate and molecular spectrum of spontaneous mutations in the bacterium Escherichia coli as determined by whole-genome sequencing. Proc. Natl Acad. Sci. USA 109, E2774–E2783 (2012).
Waine, D. J., Honeybourne, D., Smith, E. G., Whitehouse, J. L. & Dowson, C. G. Association between hypermutator phenotype, clinical variables, mucoid phenotype, and antimicrobial resistance in Pseudomonas aeruginosa. J. Clin. Microbiol. 46, 3491–3493 (2008).
Ferroni, A. et al. Effect of mutator P. aeruginosa on antibiotic resistance acquisition and respiratory function in cystic fibrosis. Pediatr. Pulmonol. 44, 820–825 (2009).
Jee, J. et al. Rates and mechanisms of bacterial mutagenesis from maximum-depth sequencing. Nature 534, 693–696 (2016).
Abascal, F. et al. Somatic mutation landscapes at single-molecule resolution. Nature 593, 405–410 (2021).
Schmitt, M. W. et al. Detection of ultra-rare mutations by next-generation sequencing. Proc. Natl Acad. Sci. USA 109, 14508–14513 (2012).
Bhawsinghka, N., Burkholder, A. & Schaaper, R. M. Detection of DNA replication errors and 8-oxo-dGTP-mediated mutations in E. coli by duplex DNA sequencing. DNA Repair 123, 103462 (2023).
Dapa, T., Ramiro, R. S., Pedro, M. F., Gordo, I. & Xavier, K. B. Diet leaves a genetic signature in a keystone member of the gut microbiota. Cell Host Microbe 30, 183–199 (2022).
Zafar, M. A., Kono, M., Wang, Y., Zangari, T. & Weiser, J. N. Infant mouse model for the study of shedding and transmission during Streptococcus pneumoniae monoinfection. Infect. Immun. 84, 2714–2722 (2016).
Vasquez, K. S. et al. Quantifying rapid bacterial evolution and transmission within the mouse intestine. Cell Host Microbe 29, 1454–1468 (2021).
Barroso-Batista, J. et al. The first steps of adaptation of Escherichia coli to the gut are dominated by soft sweeps. PLoS Genet. 10, e1004182 (2014).
Ghalayini, M. et al. Evolution of a dominant natural isolate of Escherichia coli in the human gut over the course of a year suggests a neutral evolution with reduced effective population size. Appl. Environ. Microbiol. 84, e02377-17 (2018).
Barroso-Batista, J., Demengeot, J. & Gordo, I. Adaptive immunity increases the pace and predictability of evolutionary change in commensal gut bacteria. Nat. Commun. 6, 8945 (2015).
Barreto, H. C., Sousa, A. & Gordo, I. The landscape of adaptive evolution of a gut commensal bacteria in aging mice. Curr. Biol. 30, 1102–1109 (2020).
Cowley, L. A. et al. Evolution via recombination: cell-to-cell contact facilitates larger recombination events in Streptococcus pneumoniae. PLoS Genet. 14, e1007410 (2018).
Coluzzi, C., Garcillán-Barcia, M. P., de la Cruz, F. & Rocha, E. P. C. Evolution of plasmid mobility: origin and fate of conjugative and nonconjugative plasmids. Mol. Biol. Evol. 39, msac115 (2022).
Weinert, L. A., Werren, J. H., Aebi, A., Stone, G. N. & Jiggins, F. M. Evolution and diversity of Rickettsia bacteria. BMC Biol. 7, 6 (2009).
Suchland, R. J. et al. Chromosomal recombination targets in chlamydia interspecies lateral gene transfer. J. Bacteriol. 201, e00365-19 (2019).
Chiner-Oms, Á. et al. Genomic determinants of speciation and spread of the Mycobacterium tuberculosis complex. Sci. Adv. 5, eaaw3307 (2019).
Falush, D. et al. Recombination and mutation during long-term gastric colonization by Helicobacter pylori: estimates of clock rates, recombination size, and minimal age. Proc. Natl Acad. Sci. USA 98, 15056–15061 (2001).
Nguyen, A. N. T. et al. Recombination resolves the cost of horizontal gene transfer in experimental populations of Helicobacter pylori. Proc. Natl Acad. Sci. USA 119, e2119010119 (2022).
Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012).
Conwill, A. et al. Anatomy promotes neutral coexistence of strains in the human skin microbiome. Cell Host Microbe 30, 171–182 (2022).
Shkoporov, A. N. et al. Viral biogeography of the mammalian gut and parenchymal organs. Nat. Microbiol. 7, 1301–1311 (2022).
Lourenço, M. et al. The spatial heterogeneity of the gut limits predation and fosters coexistence of bacteria and bacteriophages. Cell Host Microbe 28, 390–401 (2020).
Im, H. et al. Anatomical site-specific carbohydrate availability impacts Streptococcus pneumoniae virulence and fitness during colonization and disease. Infect. Immun. 90, e0045121 (2022).
Kaspar, J. et al. A unique open reading frame within the comX gene of Streptococcus mutans regulates genetic competence and oxidative stress tolerance. Mol. Microbiol. 96, 463–482 (2015).
Pinilla-Redondo, R., Riber, L. & Sørensen, S. J. Fluorescence recovery allows the implementation of a fluorescence reporter gene platform applicable for the detection and quantification of horizontal gene transfer in anoxic environments. Appl. Environ. Microbiol. 84, e02507-17 (2018).
Smillie, C. S. et al. Ecology drives a global network of gene exchange connecting the human microbiome. Nature 480, 241–244 (2011).
Carter, M. M. et al. Ultra-deep sequencing of Hadza hunter-gatherers recovers vanishing gut microbes. Cell 186, 3111–3124 (2023).
Groussin, M. et al. Elevated rates of horizontal gene transfer in the industrialized human microbiome. Cell 184, 2053–2067 (2021).
Sonnenburg, J. L. & Sonnenburg, E. D. Vulnerability of the industrialized microbiota. Science 366, eaaw9255 (2019).
Sheinman, M. et al. Identical sequences found in distant genomes reveal frequent horizontal transfer across the bacterial domain. eLife 10, e62719 (2021).
Redondo-Salvo, S. et al. Pathways for horizontal gene transfer in bacteria revealed by a global map of their plasmids. Nat. Commun. 11, 3602 (2020).
Hawkey, J. et al. ESBL plasmids in Klebsiella pneumoniae: diversity, transmission and contribution to infection burden in the hospital setting. Genome Med. 14, 97 (2022).
Evans, D. R. et al. Systematic detection of horizontal gene transfer across genera among multidrug-resistant bacteria in a single hospital. eLife 9, e53886 (2020).
Hall, J. P. J., Harrison, E. & Baltrus, D. A. Introduction: the secret lives of microbial mobile genetic elements. Philos. Trans. R. Soc. Lond. B 377, 20200460 (2022).
Hawlena, H., Bashey, F. & Lively, C. M. Bacteriocin-mediated interactions within and between coexisting species. Ecol. Evol. 2, 2521–2526 (2012).
Hertzog, B. B. et al. A sub-population of group A streptococcus elicits a population-wide production of bacteriocins to establish dominance in the host. Cell Host Microbe 23, 312–323 (2018).
Coyte, K. Z., Schluter, J. & Foster, K. R. The ecology of the microbiome: networks, competition, and stability. Science 350, 663–666 (2015).
Machado, D. et al. Polarization of microbial communities between competitive and cooperative metabolism. Nat. Ecol. Evol. 5, 195–203 (2021).
Stubbendieck, R. M. & Straight, P. D. Multifaceted interfaces of bacterial competition. J. Bacteriol. 198, 2145–2155 (2016).
Xavier, K. B. & Bassler, B. L. Interference with AI-2-mediated bacterial cell–cell communication. Nature 437, 750–753 (2005).
D’Souza, G. et al. Ecology and evolution of metabolic cross-feeding interactions in bacteria. Nat. Prod. Rep. 35, 455–488 (2018).
Bashey, F. Within-host competitive interactions as a mechanism for the maintenance of parasite diversity. Philos. Trans. R. Soc. Lond. B 370, 20140301 (2015).
Davies, N. G., Flasche, S., Jit, M. & Atkins, K. E. Within-host dynamics shape antibiotic resistance in commensal bacteria. Nat. Ecol. Evol. https://doi.org/10.1038/s41559-018-0786-x (2019).
Lo, S. W. et al. Pneumococcal lineages associated with serotype replacement and antibiotic resistance in childhood invasive pneumococcal disease in the post-PCV13 era: an international whole-genome sequencing study. Lancet Infect. Dis. 19, 759–769 (2019).
Croucher, N. J. et al. Population genomics of post-vaccine changes in pneumococcal epidemiology. Nat. Genet. 45, 656–663 (2013).
Croucher, N. J. et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 43, e15 (2014).
Didelot, X. & Wilson, D. J. ClonalFrameML: efficient inference of recombination in whole bacterial genomes. PLoS Comput. Biol. 11, e1004041 (2015).
Yaffe, E. & Relman, D. A. Tracking microbial evolution in the human gut using Hi-C reveals extensive horizontal gene transfer, persistence and adaptation. Nat. Microbiol. 5, 343–353 (2020).
Grodner, B. et al. Spatial mapping of mobile genetic elements and their bacterial hosts in complex microbiomes. Nat. Microbiol. https://doi.org/10.1038/s41564-024-01735-5 (2024).
Chen, L. et al. Short- and long-read metagenomics expand individualized structural variations in gut microbiomes. Nat. Commun. 13, 3175 (2022).
Pearson, T. et al. Pathogen to commensal? Longitudinal within-host population dynamics, evolution, and adaptation during a chronic >16-year Burkholderia pseudomallei infection. PLoS Pathog. 16, e1008298 (2020).
Golubchik, T. et al. Within-host evolution of Staphylococcus aureus during asymptomatic carriage. PLoS ONE 8, e61319 (2013).
Yang, Y. et al. Within-host evolution of a gut pathobiont facilitates liver translocation. Nature https://doi.org/10.1038/s41586-022-04949-x (2022).
Parkhill, J. et al. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat. Genet. 35, 32–40 (2003).
Vissa, V. D. & Brennan, P. J. The genome of Mycobacterium leprae: a minimal mycobacterial gene set. Genome Biol. 2, reviews1023.1 (2001).
Giulieri, S. G. et al. Niche-specific genome degradation and convergent evolution shaping Staphylococcus aureus adaptation during severe infections. eLife 11, e77195 (2022).
Klemm, E. J. et al. Emergence of host-adapted Salmonella Enteritidis through rapid evolution in an immunocompromised host. Nat. Microbiol. 1, 15023 (2016).
Norris, S. J. vls antigenic variation systems of Lyme disease Borrelia: eluding host immunity through both random, segmental gene conversion and framework heterogeneity. Microbiol. Spectr. https://doi.org/10.1128/microbiolspec.MDNA3-0038-2014 (2014).
Ohnishi, J., Piesman, J. & de Silva, A. M. Antigenic and genetic heterogeneity of Borrelia burgdorferi populations transmitted by ticks. Proc. Natl Acad. Sci. USA 98, 670–675 (2001).
Gatt, Y. E. & Margalit, H. Common adaptive strategies underlie within-host evolution of bacterial pathogens. Mol. Biol. Evol. 38, 1101–1121 (2021).
Lovewell, R. R. et al. Step-wise loss of bacterial flagellar torsion confers progressive phagocytic evasion. PLoS Pathog. 7, e1002253 (2011).
Martin, D. W. et al. Mechanism of conversion to mucoidy in Pseudomonas aeruginosa infecting cystic fibrosis patients. Proc. Natl Acad. Sci. USA 90, 8377–8381 (1993).
Leid, J. G. et al. The exopolysaccharide alginate protects Pseudomonas aeruginosa biofilm bacteria from IFN-gamma-mediated macrophage killing. J. Immunol. 175, 7512–7518 (2005).
Vargas, R. et al. In-host population dynamics of Mycobacterium tuberculosis complex during active disease. eLife 10, e61805 (2021).
Chen, Y. et al. Lesion heterogeneity and long-term heteroresistance in multidrug-resistant tuberculosis. J. Infect. Dis. 224, 889–893 (2021).
Nimmo, C. et al. Correction to: Whole genome sequencing Mycobacterium tuberculosis directly from sputum identifies more genetic diversity than sequencing from culture. BMC Genomics 20, 433 (2019).
Operario, D. J. et al. Prevalence and extent of heteroresistance by next generation sequencing of multidrug-resistant tuberculosis. PLoS ONE 12, e0176522 (2017).
Nimmo, C. et al. Dynamics of within-host Mycobacterium tuberculosis diversity and heteroresistance during treatment. EBioMedicine 55, 102747 (2020).
Djidjou-Demasse, R., Sofonea, M. T., Choisy, M. & Alizon, S. Within-host evolutionary dynamics of antimicrobial quantitative resistance. Math. Model. Nat. Phenom. 18, 24 (2023).
Chung, H. et al. Rapid expansion and extinction of antibiotic resistance mutations during treatment of acute bacterial respiratory infections. Nat. Commun. 13, 1231 (2022).
Vallée, M., Harding, C., Hall, J., Aldridge, P. D. & Tan, A. Exploring the in situ evolution of nitrofurantoin resistance in clinically derived uropathogenic Escherichia coli isolates. J. Antimicrob. Chemother. 78, 373–379 (2023).
Pečerska, J. et al. Quantifying transmission fitness costs of multi-drug resistant tuberculosis. Epidemics 36, 100471 (2021).
Lythgoe, K. A., Gardner, A., Pybus, O. G. & Grove, J. Short-sighted virus evolution and a germline hypothesis for chronic viral infections. Trends Microbiol. 25, 336–348 (2017).
Eckartt, K. A. et al. Compensatory evolution in NusG improves fitness of drug-resistant M. tuberculosis. Nature 628, 186–194 (2024).
Goig, G. A. et al. Effect of compensatory evolution in the emergence and transmission of rifampicin-resistant Mycobacterium tuberculosis in Cape Town, South Africa: a genomic epidemiology study. Lancet Microbe 4, e506–e515 (2023).
Loiseau, C. et al. The relative transmission fitness of multidrug-resistant Mycobacterium tuberculosis in a drug resistance hotspot. Nat. Commun. 14, 1988 (2023).
Harris, S. R. et al. Evolution of MRSA during hospital transmission and intercontinental spread. Science 327, 469–474 (2010).
Kantele, A. et al. Dynamics of intestinal multidrug-resistant bacteria colonisation contracted by visitors to a high-endemic setting: a prospective, daily, real-time sampling study. Lancet Microbe 2, e151–e158 (2021).
Hall, M., Woolhouse, M. & Rambaut, A. Epidemic reconstruction in a phylogenetics framework: transmission trees as partitions of the node set. PLoS Comput. Biol. 11, e1004613 (2015).
Wymant, C. et al. PHYLOSCANNER: inferring transmission from within- and between-host pathogen genetic diversity. Mol. Biol. Evol. https://doi.org/10.1093/molbev/msx304 (2017).
Lee, R. S., Proulx, J.-F., McIntosh, F., Behr, M. A. & Hanage, W. P. Previously undetected super-spreading of Mycobacterium tuberculosis revealed by deep sequencing. eLife 9, e53245 (2020).
Ianiro, G. et al. Variability of strain engraftment and predictability of microbiome composition after fecal microbiota transplantation across different diseases. Nat. Med. 28, 1913–1923 (2022).
Schmidt, T. S. B. et al. Drivers and determinants of strain dynamics following fecal microbiota transplantation. Nat. Med. 28, 1902–1912 (2022).
Olm, M. R. et al. inStrain profiles population microdiversity from metagenomic data and sensitively detects shared microbial strains. Nat. Biotechnol. 39, 727–736 (2021).
van Dijk, L. R. et al. StrainGE: a toolkit to track and characterize low-abundance strains in complex microbial communities. Genome Biol. 23, 74 (2022).
De Maio, N., Wu, C.-H. & Wilson, D. J. SCOTTI: efficient reconstruction of transmission within outbreaks with the structured coalescent. PLoS Comput. Biol. 12, e1005130 (2016).
Bush, S. J. et al. Genomic diversity affects the accuracy of bacterial single-nucleotide polymorphism–calling pipelines. Gigascience 9, giaa007 (2020).
Worby, C. J., Lipsitch, M. & Hanage, W. P. Within-host bacterial diversity hinders accurate reconstruction of transmission networks from genomic distance data. PLoS Comput. Biol. 10, e1003549 (2014).
Frost, S. D. W. et al. Eight challenges in phylodynamic inference. Epidemics 10, 88–92 (2015).
McCrone, J. T. & Lauring, A. S. Measurements of intrahost viral diversity are extremely sensitive to systematic errors in variant calling. J. Virol. 90, 6884–6895 (2016).
McCrone, J. T. et al. Stochastic processes constrain the within and between host evolution of influenza virus. eLife 7, e35962 (2018).
Lythgoe, K. A. et al. SARS-CoV-2 within-host diversity and transmission. Science https://doi.org/10.1126/science.abg0821 (2021).
Hall, M. D. et al. Improved characterisation of MRSA transmission using within-host bacterial sequence diversity. eLife 8, e46402 (2019).
Kono, M. et al. Single cell bottlenecks in the pathogenesis of Streptococcus pneumoniae. PLoS Pathog. 12, e1005887 (2016).
Diaz Caballero, J. et al. Mixed strain pathogen populations accelerate the evolution of antibiotic resistance in patients. Nat. Commun. 14, 4083 (2023).
Cele, S. et al. SARS-CoV-2 prolonged infection during advanced HIV disease evolves extensive immune escape. Cell Host Microbe 30, 154–162 (2022).
Scherer, E. M. et al. SARS-CoV-2 evolution and immune escape in immunocompromised patients. N. Engl. J. Med. 386, 2436–2438 (2022).
Zlitni, S. et al. Strain-resolved microbiome sequencing reveals mobile elements that drive bacterial competition on a clinical timescale. Genome Med. 12, 50 (2020).
Comas, I. et al. Whole-genome sequencing of rifampicin-resistant Mycobacterium tuberculosis strains identifies compensatory mutations in RNA polymerase genes. Nat. Genet. 44, 106–110 (2011).
Chaguza, C. et al. Within-host microevolution of Streptococcus pneumoniae is rapid and adaptive during natural colonisation. Nat. Commun. 11, 3442 (2020).
Bryant, J. M. et al. Emergence and spread of a human-transmissible multidrug-resistant nontuberculous mycobacterium. Science 354, 751–757 (2016).
Buenestado-Serrano, S. et al. Microevolution, reinfection and highly complex genomic diversity in patients with sequential isolates of Mycobacterium abscessus. Nat. Commun. 15, 2717 (2024).
Maier, L. et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature 555, 623–628 (2018).
Hall, M. B. et al. Benchmarking reveals superiority of deep learning variant callers on bacterial nanopore sequence data. eLife https://doi.org/10.7554/elife.98300.2 (2024).
Wang, B. et al. Single-cell massively-parallel multiplexed microbial sequencing (M3-seq) identifies rare bacterial populations and profiles phage infection. Nat. Microbiol. 8, 1846–1862 (2023).
McNulty, R. et al. Probe-based bacterial single-cell RNA sequencing predicts toxin regulation. Nat. Microbiol. 8, 934–945 (2023).
Ma, P. et al. Bacterial droplet-based single-cell RNA-seq reveals antibiotic-associated heterogeneous cellular states. Cell 186, 877–893 (2023).
Pensar, J. et al. Genome-wide epistasis and co-selection study using mutual information. Nucleic Acids Res. 47, e112 (2019).
Kuchina, A. et al. Microbial single-cell RNA sequencing by split-pool barcoding. Science 371, eaba5257 (2021).
Lötstedt, B., Stražar, M., Xavier, R., Regev, A. & Vickovic, S. Spatial host-microbiome sequencing reveals niches in the mouse gut. Nat. Biotechnol. 42, 1394–1403 (2024).
Didelot, X., Walker, A. S., Peto, T. E., Crook, D. W. & Wilson, D. J. Within-host evolution of bacterial pathogens. Nat. Rev. Microbiol. 14, 150–162 (2016).
Lee, K. A. et al. Cross-cohort gut microbiome associations with immune checkpoint inhibitor response in advanced melanoma. Nat. Med. 28, 535–544 (2022).
Perault, A. I. et al. Host adaptation predisposes Pseudomonas aeruginosa to type VI secretion system-mediated predation by the Burkholderia cepacia complex. Cell Host Microbe 28, 534–547 (2020).
Kerr, B., Riley, M. A., Feldman, M. W. & Bohannan, B. J. M. Local dispersal promotes biodiversity in a real-life game of rock–paper–scissors. Nature 418, 171–174 (2002).
Author information
Authors and Affiliations
Contributions
All authors contributed to the drafting, writing and editing of the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Microbiology thanks Daria Van Tyne and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tonkin-Hill, G., Ruis, C., Bentley, S.D. et al. Within-host bacterial evolution and the emergence of pathogenicity. Nat Microbiol 10, 1829–1840 (2025). https://doi.org/10.1038/s41564-025-02036-1
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41564-025-02036-1