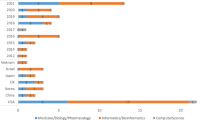
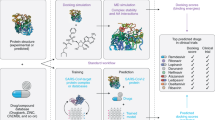
Abstract
Repurposing of existing drugs for new indications has attracted substantial attention owing to its potential to accelerate drug development and reduce costs. Hundreds of computational resources such as databases and predictive platforms have been developed that can be applied for drug repurposing, making it challenging to select the right resource for a specific drug repurposing project. With the aim of helping to address this challenge, here we overview computational approaches to drug repurposing based on a comprehensive survey of available in silico resources using a purpose-built drug repurposing ontology that classifies the resources into hierarchical categories and provides application-specific information. We also present an expert evaluation of selected resources and three drug repurposing case studies implemented within the Horizon Europe REMEDi4ALL project to demonstrate the practical use of the resources. This comprehensive Review with expert evaluations and case studies provides guidelines and recommendations on the best use of various in silico resources for drug repurposing and establishes a basis for a sustainable and extendable drug repurposing web catalogue.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Schuhmacher, A., Hinder, M., Von Stegmann Und Stein, A., Hartl, D. & Gassmann, O. Analysis of pharma R&D productivity – a new perspective needed. Drug Discov. Today 28, 103726 (2023).
Wouters, O. J., McKee, M. & Luyten, J. Estimated research and development investment needed to bring a new medicine to market, 2009–2018. JAMA 323, 844–853 (2020).
Paul, S. M. et al. How to improve R&D productivity: the pharmaceutical industry’s grand challenge. Nat. Rev. Drug Discov. 9, 203–214 (2010).
Dowden, H. & Munro, J. Trends in clinical success rates and therapeutic focus. Nat. Rev. Drug Discov. 18, 495–496 (2019).
Batta, A., Kalra, B. S. & Khirasaria, R. Trends in FDA drug approvals over last 2 decades: an observational study. J. Fam. Med. Prim. Care 9, 105–114 (2020).
Shaughnessy, A. F. Old drugs, new tricks. BMJ 342, d741 (2011).
Pushpakom, S. et al. Drug repurposing: progress, challenges and recommendations. Nat. Rev. Drug Discov. 18, 41–58 (2019).
Gupta, S. C., Sung, B., Prasad, S., Webb, L. J. & Aggarwal, B. B. Cancer drug discovery by repurposing: teaching new tricks to old dogs. Trends Pharmacol. Sci. 34, 508–517 (2013).
Austin, C. P., Mount, B. A. & Colvis, C. M. Envisioning an actionable research agenda to facilitate repurposing of off-patent drugs. Nat. Rev. Drug Discov. 20, 723–724 (2021).
Hechtelt Jonker, A. et al. IRDiRC Drug Repurposing Guidebook: making better use of existing drugs to tackle rare diseases. Nat. Rev. Drug Discov. 22, 937–938 (2023).
Zappacosta, A. R. Reversal of baldness in patient receiving minoxidil for hypertension. N. Engl. J. Med. 303, 1480–1481 (1980).
Joensuu, H. Treatment of inoperable gastrointestinal stromal tumor (GIST) with Imatinib (Glivec, Gleevec). Med. Klin. Munich Ger. 1983 97, 28–30 (2002).
Langedijk, J., Mantel-Teeuwisse, A. K., Slijkerman, D. S. & Schutjens, M.-H. D. B. Drug repositioning and repurposing: terminology and definitions in literature. Drug Discov. Today 20, 1027–1034 (2015).
Boolell, M., Gepi-Attee, S., Gingell, J. C. & Allen, M. J. Sildenafil, a novel effective oral therapy for male erectile dysfunction. Br. J. Urol. 78, 257–261 (1996).
Landgren, O. & Iskander, K. Modern multiple myeloma therapy: deep, sustained treatment response and good clinical outcomes. J. Intern. Med. 281, 365–382 (2017).
Shah, N. P. et al. Overriding imatinib resistance with a novel ABL kinase inhibitor. Science 305, 399–401 (2004).
Holderfield, M., Nagel, T. E. & Stuart, D. D. Mechanism and consequences of RAF kinase activation by small-molecule inhibitors. Br. J. Cancer 111, 640–645 (2014).
White, P. T. & Cohen, M. S. The discovery and development of sorafenib for the treatment of thyroid cancer. Expert Opin. Drug Discov. 10, 427–439 (2015).
Agrawal, K., Das, V., Vyas, P. & Hajdúch, M. Nucleosidic DNA demethylating epigenetic drugs - a comprehensive review from discovery to clinic. Pharmacol. Ther. 188, 45–79 (2018).
The RECOVERY Collaborative Group. Dexamethasone in hospitalized patients with COVID-19. N. Engl. J. Med. 384, 693–704 (2021).
Beigel, J. H. et al. Remdesivir for the treatment of COVID-19 - final report. N. Engl. J. Med. 383, 1813–1826 (2020).
Marconi, V. C. et al. Efficacy and safety of baricitinib for the treatment of hospitalised adults with COVID-19 (COV-BARRIER): a randomised, double-blind, parallel-group, placebo-controlled phase 3 trial. Lancet Respir. Med. 9, 1407–1418 (2021).
Mullard, A. FDA approves first-in-class NK3 receptor antagonist for hot flushes. Nat. Rev. Drug Discov. 22, 526 (2023).
Sahragardjoonegani, B., Beall, R. F., Kesselheim, A. S. & Hollis, A. Repurposing existing drugs for new uses: a cohort study of the frequency of FDA-granted new indication exclusivities since 1997. J. Pharm. Policy Pract. 14, 3 (2021).
Oprea, T. I. & Mestres, J. Drug repurposing: far beyond new targets for old drugs. AAPS J. 14, 759–763 (2012).
Schipper, L. J., Zeverijn, L. J., Garnett, M. J. & Voest, E. E. Can drug repurposing accelerate precision oncology? Cancer Discov. 12, 1634–1641 (2022).
Verbaanderd, C., Meheus, L., Huys, I. & Pantziarka, P. Repurposing drugs in oncology: next steps. Trends Cancer 3, 543–546 (2017).
Ji, X., Freudenberg, J. M. & Agarwal, P. Integrating biological networks for drug target prediction and prioritization. Methods Mol. Biol. 1903, 203–218 (2019).
Lin, A. et al. Off-target toxicity is a common mechanism of action of cancer drugs undergoing clinical trials. Sci. Transl. Med. 11, eaaw8412 (2019).
Wu, Y. et al. Discovery of noncancer drug effects on survival in electronic health records of patients with cancer: a new paradigm for drug repurposing. JCO Clin. Cancer Inform. 3, 1–9 (2019).
Corsello, S. M. et al. The drug repurposing hub: a next-generation drug library and information resource. Nat. Med. 23, 405–408 (2017).
Shameer, K. et al. Systematic analyses of drugs and disease indications in RepurposeDB reveal pharmacological, biological and epidemiological factors influencing drug repositioning. Brief. Bioinform. 19, 656–678 (2018).
Sam, E. & Athri, P. Web-based drug repurposing tools: a survey. Brief. Bioinform. 20, 299–316 (2019).
Song, C. M., Lim, S. J. & Tong, J. C. Recent advances in computer-aided drug design. Brief. Bioinform. 10, 579–591 (2009).
Yang, X., Wang, Y., Byrne, R., Schneider, G. & Yang, S. Concepts of artificial intelligence for computer-assisted drug discovery. Chem. Rev. 119, 10520–10594 (2019).
Tanoli, Z. et al. Exploration of databases and methods supporting drug repurposing: a comprehensive survey. Brief. Bioinform. 22, 1656–1678 (2021).
Tanoli, Z., Vähä-Koskela, M. & Aittokallio, T. Artificial intelligence, machine learning, and drug repurposing in cancer. Expert Opin. Drug Discov. 16, 977–989 (2021).
Dudley, J. T., Deshpande, T. & Butte, A. J. Exploiting drug–disease relationships for computational drug repositioning. Brief. Bioinform. 12, 303–311 (2011).
Jin, G. & Wong, S. T. C. Toward better drug repositioning: prioritizing and integrating existing methods into efficient pipelines. Drug Discov. Today 19, 637–644 (2014).
Li, J. et al. A survey of current trends in computational drug repositioning. Brief. Bioinform. 17, 2–12 (2016).
Carrasco-Ramiro, F., Peiró-Pastor, R. & Aguado, B. Human genomics projects and precision medicine. Gene Ther. 24, 551–561 (2017).
Oprea, T. I. et al. Unexplored therapeutic opportunities in the human genome. Nat. Rev. Drug Discov. 17, 317–332 (2018).
Uffelmann, E. et al. Genome-wide association studies. Nat. Rev. Methods Primers 1, 59 (2021).
Reay, W. R. & Cairns, M. J. Advancing the use of genome-wide association studies for drug repurposing. Nat. Rev. Genet. 22, 658–671 (2021).
Ochoa, D. et al. Human genetics evidence supports two-thirds of the 2021 FDA-approved drugs. Nat. Rev. Drug Discov. 21, 551–551 (2022).
Rusina, P. V. et al. Genetic support for FDA-approved drugs over the past decade. Nat. Rev. Drug Discov. 22, 864–864 (2023).
Razuvayevskaya, O., Lopez, I., Dunham, I. & Ochoa, D. Genetic factors associated with reasons for clinical trial stoppage. Nat. Genet. 56, 1862–1867 (2024).
Solomon, B. D., Nguyen, A.-D., Bear, K. A. & Wolfsberg, T. G. Clinical genomic database. Proc. Natl Acad. Sci. USA 110, 9851–9855 (2013).
Landrum, M. J. et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 42, D980–D985 (2014).
DiStefano, M. T. et al. The Gene Curation Coalition: a global effort to harmonize gene–disease evidence resources. Genet. Med. 24, 1732–1742 (2022).
Wang, Q. et al. Rare variant contribution to human disease in 281,104 UK Biobank exomes. Nature 597, 527–532 (2021).
Chen, S. et al. A genomic mutational constraint map using variation in 76,156 human genomes. Nature 625, 92–100 (2024).
Finan, C. et al. The druggable genome and support for target identification and validation in drug development. Sci. Transl. Med. 9, eaag1166 (2017).
Pinero, J. et al. DisGeNET: a discovery platform for the dynamical exploration of human diseases and their genes. Database 2015, bav028 (2015).
Bamford, S. et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br. J. Cancer 91, 355–358 (2004).
Sondka, Z. et al. COSMIC: a curated database of somatic variants and clinical data for cancer. Nucleic Acids Res. 52, D1210–D1217 (2024).
McDonagh, E. M. et al. Human genetics and genomics for drug target identification and prioritization: open targets’ perspective. Annu. Rev. Biomed. Data Sci. 7, 59–81 (2024).
Wu, P. et al. Integrating gene expression and clinical data to identify drug repurposing candidates for hyperlipidemia and hypertension. Nat. Commun. 13, 46 (2022).
Shuey, M. M. et al. A genetically supported drug repurposing pipeline for diabetes treatment using electronic health records. eBioMedicine 94, 104674 (2023).
Fernández-Torras, A., Duran-Frigola, M., Bertoni, M., Locatelli, M. & Aloy, P. Integrating and formatting biomedical data as pre-calculated knowledge graph embeddings in the Bioteque. Nat. Commun. 13, 5304 (2022).
Gordillo-Marañón, M. et al. Validation of lipid-related therapeutic targets for coronary heart disease prevention using human genetics. Nat. Commun. 12, 6120 (2021).
Ren, Y. et al. iUMRG: multi-layered network-guided propagation modeling for the inference of susceptibility genes and potential drugs against uveal melanoma. NPJ Syst. Biol. Appl. 8, 18 (2022).
Kharkar, P. S., Warrier, S. & Gaud, R. S. Reverse docking: a powerful tool for drug repositioning and drug rescue. Future Med. Chem. 6, 333–342 (2014).
Pinzi, L. & Rastelli, G. Molecular docking: shifting paradigms in drug discovery. Int. J. Mol. Sci. 20, 4331 (2019).
Lyu, J., Irwin, J. J. & Shoichet, B. K. Modeling the expansion of virtual screening libraries. Nat. Chem. Biol. 19, 712–718 (2023).
Kim, S. S., Aprahamian, M. L. & Lindert, S. Improving inverse docking target identification with Z-score selection. Chem. Biol. Drug Des. 93, 1105–1116 (2019).
Dotolo, S., Marabotti, A., Facchiano, A. & Tagliaferri, R. A review on drug repurposing applicable to COVID-19. Brief. Bioinform. 22, 726–741 (2021).
Macip, G. et al. Haste makes waste: a critical review of docking-based virtual screening in drug repurposing for SARS-CoV-2 main protease (M-pro) inhibition. Med. Res. Rev. 42, 744–769 (2022).
Pence, H. E. & Williams, A. ChemSpider: an online chemical information resource. J. Chem. Educ. 87, 1123–1124 (2010).
Irwin, J. J. et al. ZINC20—a free ultralarge-scale chemical database for ligand discovery. J. Chem. Inf. Model. 60, 6065–6073 (2020).
Berman, H. M. The Protein Data Bank. Nucleic Acids Res. 28, 235–242 (2000).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Abramson, J. et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 636, E4 (2024).
Xu, X., Huang, M. & Zou, X. Docking-based inverse virtual screening: methods, applications, and challenges. Biophys. Rep. 4, 1–16 (2018).
Gimeno, A. et al. The light and dark sides of virtual screening: what is there to know? Int. J. Mol. Sci. 20, 1375 (2019).
Morris, G. M. et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 30, 2785–2791 (2009).
Trott, O. & Olson, A. J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 (2010).
Friesner, R. A. et al. Glide: a new approach for rapid, accurate docking and scoring. 1. method and assessment of docking accuracy. J. Med. Chem. 47, 1739–1749 (2004).
Tran-Nguyen, V.-K., Junaid, M., Simeon, S. & Ballester, P. J. A practical guide to machine-learning scoring for structure-based virtual screening. Nat. Protoc. 18, 3460–3511 (2023).
Pettersen, E. F. et al. UCSF chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Seeliger, D. & De Groot, B. L. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J. Comput. Aided Mol. Des. 24, 417–422 (2010).
Rahman, N. et al. Drug repurposing for the identification of new Bcl-2 inhibitors: in vitro, STD-NMR, molecular docking, and dynamic simulation studies. Life Sci. 334, 122181 (2023).
Ajiboye, J. et al. Identification of potent and orally efficacious phosphodiesterase inhibitors in Cryptosporidium parvum-infected immunocompromised male mice. Nat. Commun. 15, 8272 (2024).
Kinnings, S. L. et al. Drug discovery using chemical systems biology: repositioning the safe medicine comtan to treat multi-drug and extensively drug resistant tuberculosis. PLoS Comput. Biol. 5, e1000423 (2009).
Salentin, S. et al. From malaria to cancer: computational drug repositioning of amodiaquine using PLIP interaction patterns. Sci. Rep. 7, 11401 (2017).
Beccari, A., Dionigi, L., Nicastri, E., Manelfi, C. & Gavioli, E. in Exscalate4CoV (eds Coletti, S. & Bernardi, G.) 19–26 (Springer, 2023); https://doi.org/10.1007/978-3-031-30691-4_3.
Carpenter, K. A. & Altman, R. B. Databases of ligand-binding pockets and protein-ligand interactions. Comput. Struct. Biotechnol. J. 23, 1320–1338 (2024).
Kim, S. et al. PubChem 2019 update: improved access to chemical data. Nucleic Acids Res. 47, D1102–D1109 (2019).
Wang, Y. et al. PubChem: a public information system for analyzing bioactivities of small molecules. Nucleic Acids Res. 37, W623–W633 (2009).
Zdrazil, B. et al. The ChEMBL database in 2023: a drug discovery platform spanning multiple bioactivity data types and time periods. Nucleic Acids Res. 52, D1180–D1192 (2024).
Omer, S. E. et al. Drug repurposing for SARS-CoV-2 main protease: molecular docking and molecular dynamics investigations. Biochem. Biophys. Rep. 29, 101225 (2022).
Mullins, J. G. L. Drug repurposing in silico screening platforms. Biochem. Soc. Trans. 50, 747–758 (2022).
Padalino, G. et al. Using ChEMBL to complement schistosome drug discovery. Pharmaceutics 15, 1359 (2023).
Milacic, M. et al. The Reactome pathway knowledgebase 2024. Nucleic Acids Res. 52, D672–D678 (2024).
Han, M., Jung, S. & Lee, D. Drug repurposing for Parkinson’s disease by biological pathway based edge-weighted network proximity analysis. Sci. Rep. 14, 21258 (2024).
Kanehisa, M., Furumichi, M., Tanabe, M., Sato, Y. & Morishima, K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 45, D353–D361 (2017).
Xu, Y., Kong, J. & Hu, P. Computational drug repurposing for Alzheimer’s disease using risk genes from GWAS and single-cell RNA sequencing studies. Front. Pharmacol. 12, 617537 (2021).
Kuleshov, M. V. et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 (2016).
Szklarczyk, D. et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 49, D605–D612 (2021).
Hermjakob, H. IntAct: an open source molecular interaction database. Nucleic Acids Res. 32, 452D–455D (2004).
Liberzon, A. et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740 (2011).
Afief, A. R. et al. Integration of genomic variants and bioinformatic-based approach to drive drug repurposing for multiple sclerosis. Biochem. Biophys. Rep. 32, 101337 (2022).
Tsherniak, A. et al. Defining a cancer dependency map. Cell 170, 564–576.e16 (2017).
Doshi, M. B. et al. Disruption of sugar nucleotide clearance is a therapeutic vulnerability of cancer cells. Nature 623, 625–632 (2023).
Bondeson, D. P. et al. Phosphate dysregulation via the XPR1–KIDINS220 protein complex is a therapeutic vulnerability in ovarian cancer. Nat. Cancer 3, 681–695 (2022).
Bi, J. et al. Targeting glioblastoma signaling and metabolism with a re-purposed brain-penetrant drug. Cell Rep. 37, 109957 (2021).
Vazquez, F. & Sellers, W. R. Are CRISPR screens providing the next generation of therapeutic targets? Cancer Res. 81, 5806–5809 (2021).
Tian, R. et al. Genome-wide CRISPRi/a screens in human neurons link lysosomal failure to ferroptosis. Nat. Neurosci. 24, 1020–1034 (2021).
Roberts, M. A. et al. Parallel CRISPR-Cas9 screens identify mechanisms of PLIN2 and lipid droplet regulation. Dev. Cell 58, 1782–1800.e10 (2023).
Corsello, S. M. et al. Discovering the anticancer potential of non-oncology drugs by systematic viability profiling. Nat. Cancer 1, 235–248 (2020).
Baranyi, M., Buday, L. & Hegedűs, B. K-Ras prenylation as a potential anticancer target. Cancer Metastasis Rev. 39, 1127–1141 (2020).
Yu, C. et al. High-throughput identification of genotype-specific cancer vulnerabilities in mixtures of barcoded tumor cell lines. Nat. Biotechnol. 34, 419–423 (2016).
Rees, M. G. et al. Systematic identification of biomarker-driven drug combinations to overcome resistance. Nat. Chem. Biol. 18, 615–624 (2022).
Payton, M. et al. Small-molecule inhibition of kinesin KIF18A reveals a mitotic vulnerability enriched in chromosomally unstable cancers. Nat. Cancer 5, 66–84 (2023).
Huang, R. et al. The NCATS pharmaceutical collection: a 10-year update. Drug Discov. Today 24, 2341–2349 (2019).
Caffall, Z. F. et al. The HIV protease inhibitor, ritonavir, corrects diverse brain phenotypes across development in mouse model of DYT-TOR1A dystonia. Sci. Transl. Med. 13, eabd3904 (2021).
Ianevski, A., Giri, A. K. & Aittokallio, T. SynergyFinder 3.0: an interactive analysis and consensus interpretation of multi-drug synergies across multiple samples. Nucleic Acids Res. 50, W739–W743 (2022).
Larsson, P. et al. Repurposing proteasome inhibitors for improved treatment of triple-negative breast cancer. Cell Death Discov. 10, 57 (2024).
Duarte, D., Rêma, A., Amorim, I. & Vale, N. Drug combinations: a new strategy to extend drug repurposing and epithelial-mesenchymal transition in breast and colon cancer cells. Biomolecules 12, 190 (2022).
Subramanian, A. et al. A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell 171, 1437–1452.e17 (2017).
Lamb, J. et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science 313, 1929–1935 (2006).
Lim, G. et al. Identification of new target proteins of a Urotensin-II receptor antagonist using transcriptome-based drug repositioning approach. Sci. Rep. 11, 17138 (2021).
Paranjpe, M. D., Taubes, A. & Sirota, M. Insights into computational drug repurposing for neurodegenerative disease. Trends Pharmacol. Sci. 40, 565–576 (2019).
Koudijs, K. K. M., Terwisscha Van Scheltinga, A. G. T., Böhringer, S., Schimmel, K. J. M. & Guchelaar, H.-J. Transcriptome signature reversion as a method to reposition drugs against cancer for precision oncology. Cancer J. 25, 116–120 (2019).
Le, B. L. et al. Transcriptomics-based drug repositioning pipeline identifies therapeutic candidates for COVID-19. Sci. Rep. 11, 12310 (2021).
Pauls, E. et al. Identification and drug-induced reversion of molecular signatures of Alzheimer’s disease onset and progression in AppNL-G-F, AppNL-F, and 3xTg-AD mouse models. Genome Med. 13, 168 (2021).
Wilk, E. J. et al. Prioritized polycystic kidney disease drug targets and repurposing candidates from pre-cystic and cystic mouse Pkd2 model gene expression reversion. Mol. Med. 29, 67 (2023).
Wang, X. et al. LINCS dataset-based repositioning of rosiglitazone as a potential anti-human adenovirus drug. Antivir. Res. 179, 104789 (2020).
Osorio, D., Tekpli, X., Kristensen, V. N. & Kuijjer, M. L. Drug combination prediction for cancer treatment using disease-specific drug response profiles and single-cell transcriptional signatures. Preprint at bioRxiv https://doi.org/10.1101/2022.03.31.486602 (2024).
Li, X. et al. The anti-leprosy drug clofazimine reduces polyQ toxicity through activation of PPARγ. eBioMedicine 103, 105124 (2024).
Carvalho, D. M. et al. Repurposing vandetanib plus everolimus for the treatment of ACVR1-mutant diffuse intrinsic pontine glioma. Cancer Discov. 12, 416–431 (2022).
Pinto, J. P., Machado, R. S. R., Xavier, J. M. & Futschik, M. E. Targeting molecular networks for drug research. Front. Genet. 5, 160 (2014).
Avram, S. et al. DrugCentral 2023 extends human clinical data and integrates veterinary drugs. Nucleic Acids Res. 51, D1276–D1287 (2023).
Davis, A. P. et al. Comparative Toxicogenomics Database (CTD): update 2023. Nucleic Acids Res. 51, D1257–D1262 (2023).
Wang, Y. et al. DrugRepo: a novel approach to repurposing drugs based on chemical and genomic features. Sci. Rep. 12, 21116 (2022).
Kuhn, M., Letunic, I., Jensen, L. J. & Bork, P. The SIDER database of drugs and side effects. Nucleic Acids Res. 44, D1075–D1079 (2016).
Tanaka, Y. et al. OnSIDES (ON-label SIDE effectS resource) database: extracting adverse drug events from drug labels using natural language processing models. Preprint at medRxiv https://doi.org/10.1101/2024.03.22.24304724 (2024).
Tatonetti, N. P., Ye, P. P., Daneshjou, R. & Altman, R. B. Data-driven prediction of drug effects and interactions. Sci. Transl. Med. 4, 125ra131 (2012).
Galeano, D., Li, S., Gerstein, M. & Paccanaro, A. Predicting the frequencies of drug side effects. Nat. Commun. 11, 4575 (2020).
Galeano, D. & Paccanaro, A. Machine learning prediction of side effects for drugs in clinical trials. Cell Rep. Methods 2, 100358 (2022).
Knox, C. et al. DrugBank 6.0: the drugbank knowledgebase for 2024. Nucleic Acids Res. 52, D1265–D1275 (2024).
Sadegh, S. et al. Network medicine for disease module identification and drug repurposing with the NeDRex platform. Nat. Commun. 12, 6848 (2021).
Williams, A. J. et al. The CompTox Chemistry Dashboard: a community data resource for environmental chemistry. J. Cheminformatics 9, 61 (2017).
Sutherland, J. J., Yonchev, D., Fekete, A. & Urban, L. A preclinical secondary pharmacology resource illuminates target-adverse drug reaction associations of marketed drugs. Nat. Commun. 14, 4323 (2023).
Nagaraj, A. B. et al. Using a novel computational drug-repositioning approach (DrugPredict) to rapidly identify potent drug candidates for cancer treatment. Oncogene 37, 403–414 (2018).
Lin, C.-Y. et al. Membrane protein-regulated networks across human cancers. Nat. Commun. 10, 3131 (2019).
Xu, C. et al. Accurate drug repositioning through non-tissue-specific core signatures from cancer transcriptomes. Cell Rep. 25, 523–535.e5 (2018).
De Bruijn, I. et al. Analysis and visualization of longitudinal genomic and clinical data from the AACR project GENIE biopharma collaborative in cbioportal. Cancer Res. 83, 3861–3867 (2023).
Variant Interpretation for Cancer Consortium. A harmonized meta-knowledgebase of clinical interpretations of somatic genomic variants in cancer. Nat. Genet. 52, 448–457 (2020).
Nikolski, M. et al. Roadmap for a European cancer data management and precision medicine infrastructure. Nat. Cancer 5, 367–372 (2024).
Ganna, A. et al. The European Health Data Space can be a boost for research beyond borders. Nat. Med. 30, 3053–3056 (2024).
Siggaard, T. et al. Disease trajectory browser for exploring temporal, population-wide disease progression patterns in 7.2 million Danish patients. Nat. Commun. 11, 4952 (2020).
Rodriguez, S. et al. Machine learning identifies candidates for drug repurposing in Alzheimer’s disease. Nat. Commun. 12, 1033 (2021).
Kc, G. B. et al. A machine learning platform to estimate anti-SARS-CoV-2 activities. Nat. Mach. Intell. 3, 527–535 (2021).
Fernández-Torras, A., Comajuncosa-Creus, A., Duran-Frigola, M. & Aloy, P. Connecting chemistry and biology through molecular descriptors. Curr. Opin. Chem. Biol. 66, 102090 (2022).
Bender, A. et al. Evaluation guidelines for machine learning tools in the chemical sciences. Nat. Rev. Chem. 6, 428–442 (2022).
Huang, K. et al. Artificial intelligence foundation for therapeutic science. Nat. Chem. Biol. 18, 1033–1036 (2022).
Wu, Z. et al. MoleculeNet: a benchmark for molecular machine learning. Chem. Sci. 9, 513–530 (2018).
Sharma, B. et al. Accurate clinical toxicity prediction using multi-task deep neural nets and contrastive molecular explanations. Sci. Rep. 13, 4908 (2023).
Barbeira, A. N. et al. Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat. Commun. 9, 1825 (2018).
Pham, T.-H., Qiu, Y., Zeng, J., Xie, L. & Zhang, P. A deep learning framework for high-throughput mechanism-driven phenotype compound screening and its application to COVID-19 drug repurposing. Nat. Mach. Intell. 3, 247–257 (2021).
Huang, K. et al. A foundation model for clinician-centered drug repurposing. Nat. Med. 30, 3601–3613 (2024).
Xu, J. et al. Interpretable deep learning translation of GWAS and multi-omics findings to identify pathobiology and drug repurposing in Alzheimer’s disease. Cell Rep. 41, 111717 (2022).
Kanemaru, K. et al. Spatially resolved multiomics of human cardiac niches. Nature 619, 801–810 (2023).
Ianevski, A. et al. Single-cell transcriptomes identify patient-tailored therapies for selective co-inhibition of cancer clones. Nat. Commun. 15, 8579 (2024).
White, J. M. et al. Drug combinations as a first line of defense against coronaviruses and other emerging viruses. mBio 12, e0334721 (2021).
Zagidullin, B. et al. DrugComb: an integrative cancer drug combination data portal. Nucleic Acids Res. 47, W43–W51 (2019).
Asano, T. et al. In silico drug screening approach using L1000-based connectivity map and its application to COVID-19. Front. Cardiovasc. Med. 9, 842641 (2022).
Bobrowski, T. et al. Synergistic and antagonistic drug combinations against SARS-CoV-2. Mol. Ther. 29, 873–885 (2021).
Liu, Y. & Zhao, H. Predicting synergistic effects between compounds through their structural similarity and effects on transcriptomes. Bioinformatics 32, 3782–3789 (2016).
Bento, A. P. et al. An open source chemical structure curation pipeline using RDKit. J. Cheminformatics 12, 51 (2020).
Jin, W. et al. Deep learning identifies synergistic drug combinations for treating COVID-19. Proc. Natl Acad. Sci. USA 118, e2105070118 (2021).
Gaulton, A. et al. The ChEMBL database in 2017. Nucleic Acids Res. 45, D945–D954 (2017).
Tanoli, Z. et al. Drug Target Commons 2.0: a community platform for systematic analysis of drug–target interaction profiles. Database 2018, 1–13 (2018).
Gilson, M. K. et al. BindingDB in 2015: a public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res. 44, D1045–D1053 (2016).
Alexander, S. P. et al. THE CONCISE GUIDE TO PHARMACOLOGY 2017/18: overview. Br. J. Pharmacol. 174, S1–S16 (2017).
Wishart, D. S. et al. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 34, D668–D672 (2006).
Wagner, A. H. et al. DGIdb 2.0: mining clinically relevant drug-gene interactions. Nucleic Acids Res. 44, D1036–D1044 (2016).
Allison, M. NCATS launches drug repurposing program. Nat. Biotechnol. 30, 571–572 (2012).
Iannelli, F. et al. Synergistic antitumor interaction of valproic acid and simvastatin sensitizes prostate cancer to docetaxel by targeting CSCs compartment via YAP inhibition. J. Exp. Clin. Cancer Res. 39, 213 (2020).
Roca, M. S. et al. Abstract 1840: repurposing of valproic acid and simvastatin in pancreatic cancer: in vitro and in vivo synergistic antitumor interaction and sensitization to gemcitabine/nab-paclitaxel via inhibition of TGFβ-EMT signaling pathway. Cancer Res. 82 (Suppl. 12), 1840 (2022).
Duran-Frigola, M. et al. Extending the small-molecule similarity principle to all levels of biology with the chemical checker. Nat. Biotechnol. 38, 1087–1096 (2020).
Zhou, Y. et al. Therapeutic target database update 2022: facilitating drug discovery with enriched comparative data of targeted agents. Nucleic Acids Res. 50, D1398–D1407 (2022).
Zhang, N., Liu, Y. & Jeong, H. Drug-drug interaction potentials of tyrosine kinase inhibitors via inhibition of UDP-glucuronosyltransferases. Sci. Rep. 5, 17778 (2015).
Goon, C. P. et al. UGT1A1 mediated drug interactions and its clinical relevance. Curr. Drug Metab. 17, 100–106 (2016).
Marques, S. C. & Ikediobi, O. N. The clinical application of UGT1A1 pharmacogenetic testing: gene-environment interactions. Hum. Genomics 4, 238–249 (2010).
Zhou, Y. et al. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 6, 14 (2020).
Cheng, F., Kovács, I. A. & Barabási, A.-L. Network-based prediction of drug combinations. Nat. Commun. 10, 1197 (2019).
Sakaeda, T., Tamon, A., Kadoyama, K. & Okuno, Y. Data mining of the public version of the FDA adverse event reporting system. Int. J. Med. Sci. 10, 796–803 (2013).
Liu, J., Lee, J., Salazar Hernandez, M. A., Mazitschek, R. & Ozcan, U. Treatment of obesity with celastrol. Cell 161, 999–1011 (2015).
Aittokallio, T. What are the current challenges for machine learning in drug discovery and repurposing? Expert Opin. Drug Discov. 17, 423–425 (2022).
Maier, A. et al. Drugst.One — a plug-and-play solution for online systems medicine and network-based drug repurposing. Nucleic Acids Res. 52, W481–W488 (2024).
Ianevski, A. et al. RepurposeDrugs: an interactive web-portal and predictive platform for repurposing mono- and combination therapies. Brief. Bioinform. 25, bbae328 (2024).
Wynants, L. et al. Prediction models for diagnosis and prognosis of covid-19: systematic review and critical appraisal. BMJ 369, m1328 (2020).
Roberts, M. et al. Common pitfalls and recommendations for using machine learning to detect and prognosticate for COVID-19 using chest radiographs and CT scans. Nat. Mach. Intell. 3, 199–217 (2021).
Sheridan, C. Can single-cell biology realize the promise of precision medicine? Nat. Biotechnol. 42, 159–162 (2024).
Pemovska, T. et al. Axitinib effectively inhibits BCR-ABL1(T315I) with a distinct binding conformation. Nature 519, 102–105 (2015).
Lee, S. et al. High-throughput identification of repurposable neuroactive drugs with potent anti-glioblastoma activity. Nat. Med. 30, 3196–3208 (2024).
Kornauth, C. et al. Functional precision medicine provides clinical benefit in advanced aggressive hematologic cancers and identifies exceptional responders. Cancer Discov. 12, 372–387 (2022).
Letai, A., Bhola, P. & Welm, A. L. Functional precision oncology: testing tumors with drugs to identify vulnerabilities and novel combinations. Cancer Cell 40, 26–35 (2022).
Dolgin, E. The future of precision cancer therapy might be to try everything. Nature 626, 470–473 (2024).
Ashburn, T. T. & Thor, K. B. Drug repositioning: identifying and developing new uses for existing drugs. Nat. Rev. Drug Discov. 3, 673–683 (2004).
Parvathaneni, V., Kulkarni, N. S., Muth, A. & Gupta, V. Drug repurposing: a promising tool to accelerate the drug discovery process. Drug Discov. Today 24, 2076–2085 (2019).
Oprea, T. I. & Overington, J. P. Computational and practical aspects of drug repositioning. Assay. Drug Dev. Technol. 13, 299–306 (2015).
Eastman, R. T. et al. Remdesivir: a review of its discovery and development leading to emergency use authorization for treatment of COVID-19. ACS Cent. Sci. 6, 672–683 (2020).
Painter, G. R., Natchus, M. G., Cohen, O., Holman, W. & Painter, W. P. Developing a direct acting, orally available antiviral agent in a pandemic: the evolution of molnupiravir as a potential treatment for COVID-19. Curr. Opin. Virol. 50, 17–22 (2021).
Huang, R. et al. Biological activity-based modeling identifies antiviral leads against SARS-CoV-2. Nat. Biotechnol. 39, 747–753 (2021).
Wang, L. et al. Susceptibility to SARS-CoV-2 of cell lines and substrates commonly used to diagnose and isolate influenza and other viruses. Emerg. Infect. Dis. 27, 1380–1392 (2021).
Morselli Gysi, D. et al. Network medicine framework for identifying drug-repurposing opportunities for COVID-19. Proc. Natl Acad. Sci. USA 118, e2025581118 (2021).
Galindez, G. et al. Lessons from the COVID-19 pandemic for advancing computational drug repurposing strategies. Nat. Comput. Sci. 1, 33–41 (2021).
Sanchez-Burgos, L., Gómez-López, G., Al-Shahrour, F. & Fernandez-Capetillo, O. An in silico analysis identifies drugs potentially modulating the cytokine storm triggered by SARS-CoV-2 infection. Sci. Rep. 12, 1626 (2022).
Lobo, S. M. et al. Efficacy of oral 20-hydroxyecdysone (BIO101), a MAS receptor activator, in adults with severe COVID-19 (COVA): a randomized, placebo-controlled, phase 2/3 trial. eClinicalMedicine 68, 102383 (2024).
Brimacombe, K. R. et al. An OpenData portal to share COVID-19 drug repurposing data in real time. Preprint at bioRxiv https://doi.org/10.1101/2020.06.04.135046 (2020).
Acknowledgements
The authors thank authors of the resources who have made their data and tools publicly available. They thank J. Tang for his valuable suggestions when conducting the survey. This work was supported by the REMEDi4ALL project, which has received funding from the European Union’s Horizon Europe research and innovation programme under grant agreement No. 101057442. Views and opinions expressed are those of the authors only and do not necessarily reflect those of the European Union, who cannot be held responsible for them. Other funding sources include the Research Council of Finland (grant 351507 to Z.T. and grants 345803 and 340141 to T.A.) and Cancer Society of Finland (grants 4709137 and 4706788 to T.A.). For questions about the web catalogue, resource annotations, expert survey or the user guide, please contact Z.T. (ziaurrehman.tanoli@helsinki.fi).
Author information
Authors and Affiliations
Contributions
Development of the repurposing ontology: Z.T. Implementation of the online catalogue website: A.K., Z.T., T.A. Data analysis: Z.T, A.F.-T., U.O.Ö., L.F., Y.G., R.G.-S., J.C.-P., T.A. Demonstrator use cases: Z.T., A.F.-T., A.B., O.S., P.G., D.C.L., Y.G., P.Ö., J.Q., J.C.-P., M.F., L.F., B.S.-L., M.T., A.K., T.A. User guide for the top-15 resources: Z.T., K.M.N., U.O.Ö., L.F., M.V.-K., U.S., M.M., A.I., T.A. Annotations of the resources in the web catalogue: Z.T., M.V.-K., U.S., M.F., J.Q., T.A. Evaluation of the resources: Z.T., A.F.-T., M.V.-K., M.d.K., A.I., Y.G., P.G., O.S., U.S., H.L., M.F., L.F., J.Q., J.C.-P., T.A., M.M., K.W., H.X., A.E.U., B.S.-L., W.S., F.B., E.B., S.P., J.S., A.P., M.J., S.C., I.G.G., T.C., A.R.B. Leading the survey implementation: Z.T. and T.A. Figures: Z.T., A.F.-T., U.O.Ö., K.M.N. Drafting the manuscript: Z.T., A.F.-T., M.F., J.Q., J.C.-P., M.d.K., K.W., T.A. All the authors read and approved the final version.
Corresponding authors
Ethics declarations
Competing interests
A.F.-T., I.G.G., R.G.-S., J.Q. and J.M. are employees of Chemotargets, a partner of the REMEDi4ALL project. ClarityVista is a software platform developed by Chemotargets. J.C.-P. and O.S. declare ownership in Phenaros Pharmaceuticals AB. T.C. and A.R.B. are employees of Dompé farmaceutici SpA, a partner of the REMEDi4ALL project. All the other authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Drug Discovery thanks David Ochoa and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
ChatGPT-4: https://openai.com/index/gpt-4/
ClinGen: https://clinicalgenome.org
ClinicalTrials: clinicaltrials.gov
Cure ID: https://cure.ncats.io/home
Drug Interaction Checker: https://go.drugbank.com/drug-interaction-checker
Drug Repurposing Central: https://drugrepocentral.scienceopen.com/collection/fee38437-df5d-4641-bd7f-b1b61882cd75
FinnGen: https://www.finngen.fi/en
Genebass: https://app.genebass.org
GTEx: https://gtexportal.org/home/
Human Cell Atlas: https://www.humancellatlas.org
In-silico Drug Repurposing Catalogue - REMEDi4ALL: https://remedi4all.org/in-silico-drug-repurposing-catalogue/
Orphanet: https://www.orpha.net
PheWAS Portal: https://azphewas.com
PubMed: https://pubmed.ncbi.nlm.nih.gov/
REMEDi4ALL project: https://remedi4all.org/
Sage Bionetworks: https://sagebionetworks.org
UK Biobank: https://www.ukbiobank.ac.uk
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tanoli, Z., Fernández-Torras, A., Özcan, U.O. et al. Computational drug repurposing: approaches, evaluation of in silico resources and case studies. Nat Rev Drug Discov 24, 521–542 (2025). https://doi.org/10.1038/s41573-025-01164-x
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41573-025-01164-x
This article is cited by
-
Identification of natural product inhibitors as potential drug candidates for treating Alzheimer's disease: molecular docking, molecular dynamics simulations, MM/GBSA and pharmacokinetics
Network Modeling Analysis in Health Informatics and Bioinformatics (2025)