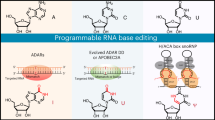
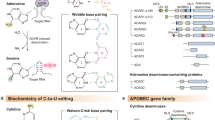
Abstract
The AID/APOBEC polynucleotide cytidine deaminases have historically been classified as either DNA mutators or RNA editors based on their first identified nucleic acid substrate preference. DNA mutators can generate functional diversity at antibody genes but also cause genomic instability in cancer. RNA editors can generate informational diversity in the transcriptome of innate immune cells, and of cancer cells. Members of both classes can act as antiviral restriction factors. Recent structural work has illuminated differences and similarities between AID/APOBEC enzymes that can catalyse DNA mutation, RNA editing or both, suggesting that the strict functional classification of members of this family should be reconsidered. As many of these enzymes have been employed for targeted genome (or transcriptome) editing, a more holistic understanding will help improve the design of therapeutically relevant programmable base editors.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Crick, F. H. On protein synthesis. Symposia Soc. Exp. Biol. 12, 138–163 (1958).
Blencowe, B. J. Alternative splicing: new insights from global analyses. Cell 126, 37–47 (2006).
Di Giammartino, D. C., Nishida, K. & Manley, J. L. Mechanisms and consequences of alternative polyadenylation. Mol. Cell 43, 853–866 (2011).
Ryvkin, P. et al. HAMR: high-throughput annotation of modified ribonucleotides. RNA 19, 1684–1692 (2013).
Helm, M. & Motorin, Y. Detecting RNA modifications in the epitranscriptome: predict and validate. Nat. Rev. Genet. 18, 275–291 (2017).
Benne, R. et al. Major transcript of the frameshifted coxll gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell 46, 819–826 (1986).
Bass, B. L. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 71, 817–846 (2002).
Nishikura, K. Functions and Regulation of RNA Editing by ADAR Deaminases. Annu. Rev. Biochem. 79, 321–349 (2010).
Savva, Y. A., Rieder, L. E. & Reenan, R. A. The ADAR protein family. Genome Biol. 13, 252 (2012).
Wedekind, J. E., Dance, G. S. C., Sowden, M. P. & Smith, H. C. Messenger RNA editing in mammals: new members of the APOBEC family seeking roles in the family business. Trends Genet. 19, 207–216 (2003).
Lorenzo, J. P. et al. APOBEC2 is a transcriptional repressor required for proper myoblast differentiation. Preprint at bioRxiv https://doi.org/10.1101/2020.07.29.223594 (2021).
Reardon, S. Step aside CRISPR, RNA editing is taking off. Nature 578, 24–27 (2020).
Salter, J. D. & Smith, H. C. Modeling the embrace of a mutator: APOBEC selection of nucleic acid ligands. Trends Biochem. Sci. 43, 606–622 (2018).
Salter, J. D., Bennett, R. P. & Smith, H. C. The APOBEC protein family: united by structure, divergent in function. Trends Biochemical Sci. 41, 578–594 (2016).
Bohn, J. A. et al. APOBEC3H structure reveals an unusual mechanism of interaction with duplex RNA. Nat. Commun. 8, 1021 (2017).
Kouno, T. et al. Crystal structure of APOBEC3A bound to single-stranded DNA reveals structural basis for cytidine deamination and specificity. Nat. Commun. 8, 15024 (2017).
Matsuoka, T. et al. Structural basis of chimpanzee APOBEC3H dimerization stabilized by double-stranded RNA. Nucleic Acids Res. 46, 10368–10379 (2018).
Rathore, A. et al. The local dinucleotide preference of APOBEC3G can be altered from 5′-CC to 5′-TC by a single amino acid substitution. J. Mol. Biol. 425, 4442–4454 (2013).
Shaban, N. M. et al. The antiviral and cancer genomic DNA deaminase APOBEC3H is regulated by an RNA-mediated dimerization mechanism. Mol. Cell 69, 75–86.e9 (2018).
Shi, K. et al. Structural basis for targeted DNA cytosine deamination and mutagenesis by APOBEC3A and APOBEC3B. Nat. Struct. Mol. Biol. 24, 131–139 (2017).
Wolfe, A. D., Li, S., Goedderz, C. & Chen, X. S. The structure of APOBEC1 and insights into its RNA and DNA substrate selectivity. NAR Cancer 2, zcaa027 (2020).
Maiti, A. et al. Crystal structure of the catalytic domain of HIV-1 restriction factor APOBEC3G in complex with ssDNA. Nat. Commun. 9, 2460 (2018).
Polevoda, B. et al. DNA mutagenic activity and capacity for HIV-1 restriction of the cytidine deaminase APOBEC3G depend on whether DNA or RNA binds to tyrosine 315. J. Biol. Chem. 292, 8642–8656 (2017).
King, J. J. & Larijani, M. A novel regulator of activation-induced cytidine deaminase/APOBECs in immunity and cancer: Schrödinger’s CATalytic Pocket. Front. Immunol. 8, 351 (2017).
Olson, M. E., Harris, R. S. & Harki, D. A. APOBEC enzymes as targets for virus and cancer therapy. Cell Chem. Biol. 25, 36–49 (2018).
Buisson, R. et al. Passenger hotspot mutations in cancer driven by APOBEC3A and mesoscale genomic features. Science 364, eaaw2872 (2019).
Holtz, C. M., Sadler, H. A. & Mansky, L. M. APOBEC3G cytosine deamination hotspots are defined by both sequence context and single-stranded DNA secondary structure. Nucleic Acids Res. 41, 6139–6148 (2013).
Jalili, P. et al. Quantification of ongoing APOBEC3A activity in tumor cells by monitoring RNA editing at hotspots. Nat. Commun. 11, 2971 (2020).
Sharma, S. & Baysal, B. E. Stem–loop structure preference for site-specific RNA editing by APOBEC3A and APOBEC3G. PeerJ 5, e4136 (2017).
Maris, C., Masse, J., Chester, A. N. N., Navaratnam, N. & Allain, F. H. NMR structure of the apoB mRNA stem−loop and its interaction with the C to U editing APOBEC1 complementary factor. RNA 11, 173–186 (2005).
Richardson, N., Navaratnam, N. & Scott, J. Secondary structure for the apolipoprotein B mRNA editing site. J. Biol. Chem. 273, 31707–31717 (1998).
Shah, R. R. et al. Sequence requirements for the editing of apolipoprotein B mRNA. J. Biol. Chem. 266, 16301–16304 (1991).
Losey, H. C., Ruthenburg, A. J. & Verdine, G. L. Crystal structure of Staphylococcus aureus tRNA adenosine deaminase TadA in complex with RNA. Nat. Struct. Mol. Biol. 13, 153–159 (2006).
Ito, F. et al. Understanding the structure, multimerization, subcellular localization and mC selectivity of a genomic mutator and anti-HIV factor APOBEC3H. Sci. Rep. 8, 3763 (2018).
Mitra, M. et al. Structural determinants of human APOBEC3A enzymatic and nucleic acid binding properties. Nucleic Acids Res. 42, 1095–1110 (2014).
Tang, G. et al. Creating RNA specific C-to-U editase from APOBEC3A by separation of its activities on DNA and RNA substrates. ACS Synth. Biol. 10, 1106–1115 (2021).
Qiao, Q. et al. AID recognizes structured DNA for class switch recombination. Mol. Cell 67, 361–373.e4 (2017).
Devos, J. M., Tomanicek, S. J., Jones, C. E., Nossal, N. G. & Mueser, T. C. Crystal structure of bacteriophage T4 5′ nuclease in complex with a branched DNA reveals how flap endonuclease-1 family nucleases bind their substrates. J. Biol. Chem. 282, 31713–31724 (2007).
Jiang, F. et al. Structures of a CRISPR–Cas9 R-loop complex primed for DNA cleavage. Science 351, 867–871 (2016).
Zheng, S. et al. Non-coding RNA generated following lariat debranching mediates targeting of AID to DNA. Cell 161, 762–773 (2015).
Dickerson, S. K., Market, E., Besmer, E. & Papavasiliou, F. N. AID mediates hypermutation by deaminating single stranded DNA. J. Exp. Med. 197, 1291–1296 (2003).
Abdouni, H. S. et al. DNA/RNA hybrid substrates modulate the catalytic activity of purified AID. Mol. Immunol. 93, 94–106 (2018).
Fritz, E. L. et al. A comprehensive analysis of the effects of the deaminase AID on the transcriptome and methylome of activated B cells. Nat. Immunol. 14, 749–755 (2013).
Krzysiak, T. C., Jung, J., Thompson, J., Baker, D. & Gronenborn, A. M. APOBEC2 is a monomer in solution: implications for APOBEC3G models. Biochemistry 51, 2008–2017 (2012).
Prochnow, C., Bransteitter, R., Klein, M. G., Goodman, M. F. & Chen, X. S. The APOBEC-2 crystal structure and functional implications for the deaminase AID. Nature 445, 447–451 (2007).
Ataie, N. J. et al. Zinc coordination geometry and ligand binding affinity: the structural and kinetic analysis of the second-shell serine 228 residue and the methionine 180 residue of the aminopeptidase from Vibrio proteolyticus. Biochemistry 47, 7673–7683 (2008).
Boyaci, H., Chen, J., Jansen, R., Darst, S. A. & Campbell, E. A. Structures of an RNA polymerase promoter melting intermediate elucidate DNA unwinding. Nature 565, 382–385 (2019).
Powell, C., Cornblath, E. & Goldman, D. Zinc-binding domain-dependent, deaminase-independent actions of apolipoprotein B mRNA-editing enzyme, catalytic polypeptide 2 (Apobec2), mediate its effect on zebrafish retina regeneration. J. Biol. Chem. 289, 28924–28941 (2014).
Chester, A. et al. The apolipoprotein B mRNA editing complex performs a multifunctional cycle and suppresses nonsense-mediated decay. EMBO J. 22, 3971–3982 (2003).
Ito, S. et al. Activation-induced cytidine deaminase shuttles between nucleus and cytoplasm like apolipoprotein B mRNA editing catalytic polypeptide 1. Proc. Natl Acad. Sci. USA 101, 1975–1980 (2004).
Patenaude, A.-M. et al. Active nuclear import and cytoplasmic retention of activation-induced deaminase. Nat. Struct. Mol. Biol. 16, 517–527 (2009).
Lackey, L., Law, E. K., Brown, W. L. & Harris, R. S. Subcellular localization of the APOBEC3 proteins during mitosis and implications for genomic DNA deamination. Cell Cycle 12, 762–772 (2013).
Salamango, D. J. et al. APOBEC3H subcellular localization determinants define zipcode for targeting HIV-1 for restriction. Mol. Cell Biol. 38, e00356–18 (2018).
Bennett, R. P. et al. APOBEC-1 and AID are nucleo-cytoplasmic trafficking proteins but APOBEC3G cannot traffic. Biochem. Biophys. Res. Commun. 350, 214–219 (2006).
Bennett, R. P., Presnyak, V., Wedekind, J. E. & Smith, H. C. Nuclear exclusion of the HIV-1 host defense factor APOBEC3G requires a novel cytoplasmic retention signal and is not dependent on RNA binding. J. Biol. Chem. 283, 7320–7327 (2008).
Navaratnam, N. et al. The p27 catalytic subunit of the apolipoprotein B mRNA editing enzyme is a cytidine deaminase. J. Biol. Chem. 268, 20709–20712 (1993).
Teng, B. B., Burant, C. F. & Davidson, N. O. Molecular cloning of an apolipoprotein B messenger RNA editing protein. Science 260, 1816–1819 (1993).
Lellek, H. et al. Purification and molecular cloning of a novel essential component of the apolipoprotein B mRNA editing enzyme-complex. J. Biol. Chem. 275, 19848–19856 (2000).
Mehta, A., Kinter, M. T., Sherman, N. E. & Driscoll, D. M. Molecular cloning of apobec-1 complementation factor, a novel RNA-binding protein involved in the editing of apolipoprotein B mRNA. Mol. Cell. Biol. 20, 1846–1854 (2000).
Fossat, N. et al. C to U RNA editing mediated by APOBEC1 requires RNA-binding protein RBM47. EMBO Rep 15, 903–910 (2014).
Blanc, V. et al. Apobec1 complementation factor (A1CF) and RBM47 interact in tissue-specific regulation of C to U RNA editing in mouse intestine and liver. RNA 25, 70–81 (2019).
Soleymanjahi, S., Blanc, V. & Davidson, N. APOBEC1 mediated C-to-U RNA editing: target sequence and trans-acting factor contribution to 177 RNA editing events in 119 murine transcripts in-vivo. RNA 27, 876–890 (2021).
Rayon-Estrada, V. et al. Epitranscriptomic profiling across cell types reveals associations between APOBEC1-mediated RNA editing, gene expression outcomes, and cellular function. Proc. Natl Acad. Sci. USA 114, 13296–13301 (2017).
Lerner, T., Papavasiliou, F. N. & Pecori, R. RNA editors, cofactors, and mRNA targets: an overview of the C-to-U RNA editing machinery and its implication in human disease. Genes 10, 13 (2018).
Basu, U. et al. The AID antibody diversification enzyme is regulated by protein kinase A phosphorylation. Nature 438, 508–511 (2005).
Chaudhuri, J. & Alt, F. W. Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nat. Rev. Immunol. 4, 541–552 (2004).
Conticello, S. G. et al. Interaction between antibody-diversification enzyme AID and spliceosome-associated factor CTNNBL1. Mol. Cell 31, 474–484 (2008).
McBride, K. M. et al. Regulation of hypermutation by activation-induced cytidine deaminase phosphorylation. Biochem. Soc. Trans. 103, 8798–8803 (2006).
Pasqualucci, L., Kitaura, Y., Gu, H. & Dalla-Favera, R. PKA-mediated phosphorylation regulates the function of activation-induced deaminase (AID) in B cells. Proc. Natl Acad. Sci. USA 103, 395–400 (2006).
Vuong, B. Q. et al. Specific recruitment of protein kinase A to the immunoglobulin locus regulates class-switch recombination. Nat. Immunol. 10, 420–426 (2009).
Marx, A., Galilee, M. & Alian, A. Zinc enhancement of cytidine deaminase activity highlights a potential allosteric role of loop-3 in regulating APOBEC3 enzymes. Sci. Rep. 5, 18191 (2015).
Chen, J. & MacCarthy, T. The preferred nucleotide contexts of the AID/APOBEC cytidine deaminases have differential effects when mutating retrotransposon and virus sequences compared to host genes. PLOS Comput. Biol. 13, e1005471 (2017).
Eisenberg, E. & Levanon, E. Y. A-to-I RNA editing — immune protector and transcriptome diversifier. Nat. Rev. Genet. 19, 473–490 (2018).
Martinez, T., Shapiro, M., Bhaduri-McIntosh, S. & MacCarthy, T. Evolutionary effects of the AID/APOBEC family of mutagenic enzymes on human gamma-herpesviruses. Virus Evol. 5, vey040 (2019).
Madani, N. & Kabat, D. An endogenous inhibitor of human immunodeficiency virus in human lymphocytes is overcome by the viral Vif protein. J. Virol. 72, 10251–10255 (1998).
Simon, J. H., Gaddis, N. C., Fouchier, R. A. & Malim, M. H. Evidence for a newly discovered cellular anti-HIV-1 phenotype. Nat. Med. 4, 1397–1400 (1998).
Harris, R. S. et al. DNA deamination mediates innate immunity to retroviral infection. Cell 113, 803–809 (2003).
Mangeat, B. et al. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424, 99–103 (2003).
Sheehy, A. M., Gaddis, N. C., Choi, J. D. & Malim, M. H. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418, 646–650 (2002).
Dang, Y. et al. Human cytidine deaminase APOBEC3H restricts HIV-1 replication. J. Biol. Chem. 283, 11606–11614 (2008).
Harris, R. S. & Dudley, J. P. APOBECs and virus restriction. Virology 479–480, 131–145 (2015).
Hayward, J. A. et al. Differential evolution of antiretroviral restriction factors in pteropid bats as revealed by APOBEC3 gene complexity. Mol. Biol. Evol. 35, 1626–1637 (2018).
Liddament, M. T., Brown, W. L., Schumacher, A. J. & Harris, R. S. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr. Biol. 14, 1385–1391 (2004).
Domingo, E., Sheldon, J. & Perales, C. Viral quasispecies evolution. Microbiol. Mol. Biol. Rev. 76, 159–216 (2012).
Kim, E.-Y. et al. Human APOBEC3G-mediated editing can promote HIV-1 sequence diversification and accelerate adaptation to selective pressure. J. Virol. 84, 10402–10405 (2010).
Wood, N. et al. HIV evolution in early infection: selection pressures, patterns of insertion and deletion, and the impact of APOBEC. PLoS Pathog. 5, e1000414 (2009).
Venkatesan, S. et al. Perspective: APOBEC mutagenesis in drug resistance and immune escape in HIV and cancer evolution. Ann. Oncol. 29, 563–572 (2018).
Bonvin, M. et al. Interferon-inducible expression of APOBEC3 editing enzymes in human hepatocytes and inhibition of hepatitis B virus replication. Hepatology 43, 1364–1374 (2006).
Bulliard, Y. et al. Structure–function analyses point to a polynucleotide-accommodating groove essential for APOBEC3A restriction activities. J. Virol. 85, 1765–1776 (2011).
Warren, C. J. et al. APOBEC3A functions as a restriction factor of human papillomavirus. J. Virol. 89, 688–702 (2014).
Di Giorgio, S., Martignano, F., Torcia, M. G., Mattiuz, G. & Conticello, S. G. Evidence for host-dependent RNA editing in the transcriptome of SARS-CoV-2. Sci. Adv. 6, eabb5813 (2020).
Graudenzi, A., Maspero, D., Angaroni, F., Piazza, R. & Ramazzotti, D. Mutational signatures and heterogeneous host response revealed via large-scale characterization of SARS-CoV-2 genomic diversity. iScience 24, 102116 (2021).
Picardi, E., Mansi, L. & Pesole, G. Detection of A-to-I RNA editing in SARS-COV-2. Genes 13, 41 (2021).
Simmonds, P. & Ansari, M. A. Extensive C->U transition biases in the genomes of a wide range of mammalian RNA viruses; potential associations with transcriptional mutations, damage- or host-mediated editing of viral RNA. PLoS Pathog. 17, e1009596 (2021).
Kim, K. et al. APOBEC-mediated editing of SARS-CoV-2 genomic RNA impacts viral replication and fitness. Preprint at bioRxiv https://www.biorxiv.org/content/10.1101/2021.12.18.473309v1 (2021).
Klimczak, L. J., Randall, T. A., Saini, N., Li, J.-L. & Gordenin, D. A. Similarity between mutation spectra in hypermutated genomes of rubella virus and in SARS-CoV-2 genomes accumulated during the COVID-19 pandemic. PLoS ONE 15, e0237689 (2020).
Blanco-Melo, D. et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell 181, 1036–1045.e9 (2020).
Cotroneo, C. E., Mangano, N., Dragani, T. A. & Colombo, F. Lung expression of genes putatively involved in SARS-CoV-2 infection is modulated in cis by germline variants. Eur. J. Hum. Genet. 29, 1019–1026 (2021).
Liao, M. et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 26, 842–844 (2020).
Mourier, T. et al. Host-directed editing of the SARS-CoV-2 genome. Biochem. Biophys. Res. Commun. 538, 35–39 (2021).
Ratcliff, J. & Simmonds, P. Potential APOBEC-mediated RNA editing of the genomes of SARS-CoV-2 and other coronaviruses and its impact on their longer term evolution. Virology 556, 62–72 (2021).
van Dorp, L. et al. Emergence of genomic diversity and recurrent mutations in SARS-CoV-2. Infect. Genet. Evol. 83, 104351 (2020).
Matyášek, R. & Kovařík, A. Mutation patterns of human SARS-CoV-2 and Bat RaTG13 coronavirus genomes are strongly biased towards C > U transitions, indicating rapid evolution in their hosts. Genes 11, E761 (2020).
Wang, R., Hozumi, Y., Zheng, Y.-H., Yin, C. & Wei, G.-W. Host immune response driving SARS-CoV-2 evolution. Viruses 12, 1095 (2020).
Milewska, A. et al. APOBEC3-mediated restriction of RNA virus replication. Sci. Rep. 8, 5960 (2018).
Yamanaka, S. et al. Apolipoprotein B mRNA-editing protein induces hepatocellular carcinoma and dysplasia in transgenic animals. Proc. Natl Acad. Sci. USA 92, 8483–8487 (1995).
Franco, S. et al. H2AX prevents DNA breaks from progressing to chromosome breaks and translocations. Mol. Cell 21, 201–214 (2006).
Ramiro, A. R. et al. Role of genomic instability and p53 in AID-induced c-myc–Igh translocations. Nature 440, 105–109 (2006).
Harris, R. S., Petersen-Mahrt, S. K. & Neuberger, M. S. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol. Cell 10, 1247–1253 (2002).
Landry, S., Narvaiza, I., Linfesty, D. C. & Weitzman, M. D. APOBEC3A can activate the DNA damage response and cause cell-cycle arrest. EMBO Rep. 12, 444–450 (2011).
Alexandrov, L. B. et al. The repertoire of mutational signatures in human cancer. Nature 578, 94–101 (2020).
Roberts, S. A. et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat. Genet. 45, 970–976 (2013).
Nik-Zainal, S. et al. Mutational processes molding the genomes of 21 breast cancers. Cell 149, 979–993 (2012).
Roberts, S. A. et al. Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions. Mol. Cell 46, 424–435 (2012).
Chan, K. & Gordenin, D. A. Clusters of multiple mutations: incidence and molecular mechanisms. Annu. Rev. Genet. 49, 243–267 (2015).
ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Pan-cancer analysis of whole genomes. Nature 578, 82–93 (2020).
Alexandrov, L. B. et al. Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013).
Klemm, L. et al. The B cell mutator AID promotes B lymphoid blast crisis and drug resistance in chronic myeloid leukemia. Cancer Cell 16, 232–245 (2009).
Law, E. K. et al. The DNA cytosine deaminase APOBEC3B promotes tamoxifen resistance in ER-positive breast cancer. Sci. Adv. 2, e1601737 (2016).
de Bruin, E. C. et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science 346, 251–256 (2014).
Venkatesan, S. et al. Induction of APOBEC3 exacerbates DNA replication stress and chromosomal instability in early breast and lung cancer evolution. Cancer Discov. 11, 2456–2473 (2021).
Cahill, D. P., Kinzler, K. W., Vogelstein, B. & Lengauer, C. Genetic instability and darwinian selection in tumours. Trends Cell Biol. 9, M57–M60 (1999).
Weaver, B. A. A., Silk, A. D., Montagna, C., Verdier-Pinard, P. & Cleveland, D. W. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell 11, 25–36 (2007).
Swanton, C., McGranahan, N., Starrett, G. J. & Harris, R. S. APOBEC enzymes: mutagenic fuel for cancer evolution and heterogeneity. Cancer Discov. 5, 704–712 (2015).
Serebrenik, A. A. et al. The deaminase APOBEC3B triggers the death of cells lacking uracil DNA glycosylase. Proc. Natl Acad. Sci. USA 116, 22158–22163 (2019).
Asaoka, M., Ishikawa, T., Takabe, K. & Patnaik, S. K. APOBEC3-mediated RNA editing in breast cancer is associated with heightened immune activity and improved survival. Int. J. Mol. Sci. 20, 5621 (2019).
Ben-Aroya, S. & Levanon, E. Y. A-to-I RNA editing: an overlooked source of cancer mutations. Cancer Cell 33, 789–790 (2018).
Christofi, T. & Zaravinos, A. RNA editing in the forefront of epitranscriptomics and human health. J. Transl. Med. 17, 319 (2019).
Driscoll, C. B. et al. APOBEC3B-mediated corruption of the tumor cell immunopeptidome induces heteroclitic neoepitopes for cancer immunotherapy. Nat. Commun. 11, 790 (2020).
Paz-Yaacov, N. et al. Elevated RNA editing activity is a major contributor to transcriptomic diversity in tumors. Cell Rep. 13, 267–276 (2015).
Ramírez-Moya, J., Baker, A. R., Slack, F. J. & Santisteban, P. ADAR1-mediated RNA editing is a novel oncogenic process in thyroid cancer and regulates miR-200 activity. Oncogene 39, 3738–3753 (2020).
Blanc, V. et al. Deletion of the AU-rich RNA binding protein apobec-1 reduces intestinal tumor burden in Apcmin mice. Cancer Res. 67, 8565–8573 (2007).
Nelson, V. R., Heaney, J. D., Tesar, P. J., Davidson, N. O. & Nadeau, J. H. Transgenerational epigenetic effects of the Apobec1 cytidine deaminase deficiency on testicular germ cell tumor susceptibility and embryonic viability. Proc. Natl Acad. Sci. USA 109, E2766–E2773 (2012).
Casati, B., Stamkopoulou, D., Tasakis, R. N. & Pecori, R. in Epitranscriptomics (eds Jurga, S. & Barciszewski, J.) 471–503 (Springer International, 2021).
Khosravi, H. M. & Jantsch, M. F. Site-directed RNA editing: recent advances and open challenges. RNA Biol. 18, 41–50 (2021).
Montiel-Gonzalez, M. F., Diaz Quiroz, J. F. & Rosenthal, J. J. C. Current strategies for site-directed RNA editing using ADARs. Methods 156, 16–24 (2019).
Park, S. & Beal, P. A. Off-target editing by CRISPR-guided DNA base editors. Biochemistry 58, 3727–3734 (2019).
Rees, H. A. & Liu, D. R. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 19, 770–788 (2018).
Vogel, P. & Stafforst, T. Critical review on engineering deaminases for site-directed RNA editing. Curr. Opin. Biotechnol. 55, 74–80 (2018).
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A. & Liu, D. R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016).
Petersen-Mahrt, S. K. & Neuberger, M. S. In vitro deamination of cytosine to uracil in single-stranded DNA by apolipoprotein B editing complex catalytic subunit 1 (APOBEC1). J. Biol. Chem. 278, 19583–19586 (2003).
Saraconi, G., Severi, F., Sala, C., Mattiuz, G. & Conticello, S. G. The RNA editing enzyme APOBEC1 induces somatic mutations and a compatible mutational signature is present in esophageal adenocarcinomas. Genome Biol. 15, 417 (2014).
Grünewald, J. et al. Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base editors. Nature 569, 433–437 (2019).
Zhou, C. et al. Off-target RNA mutation induced by DNA base editing and its elimination by mutagenesis. Nature 571, 275–278 (2019).
Abudayyeh, O. O. et al. A cytosine deaminase for programmable single-base RNA editing. Science 365, 382–386 (2019).
Hersberger, M., Patarroyo-white, S., Arnold, K. S. & Innerarity, T. L. Phylogenetic analysis of the apolipoprotein B mRNA-editing region. J. Biol. Chem. 274, 34590–34597 (1999).
Huang, X. et al. Programmable C-to-U RNA editing using the human APOBEC3A deaminase. EMBO J. 40, e108209 (2021).
Bhakta, S., Sakari, M. & Tsukahara, T. RNA editing of BFP, a point mutant of GFP, using artificial APOBEC1 deaminase to restore the genetic code. Sci. Rep. 10, 17304 (2020).
Stroppel, A. S. et al. Harnessing self-labeling enzymes for selective and concurrent A-to-I and C-to-U RNA base editing. Nucleic Acids Res. 49, e95 (2021).
Vogel, P. et al. Efficient and precise editing of endogenous transcripts with SNAP-tagged ADARs. Nat. Methods 15, 535–538 (2018).
Merkle, T. et al. Precise RNA editing by recruiting endogenous ADARs with antisense oligonucleotides. Nat. Biotechnol. 37, 133–138 (2019).
Qu, L. et al. Programmable RNA editing by recruiting endogenous ADAR using engineered RNAs. Nat. Biotechnol. 37, 1059–1069 (2019).
Liew, Y. J., Li, Y., Baumgarten, S., Voolstra, C. R. & Aranda, M. Condition-specific RNA editing in the coral symbiont Symbiodinium microadriaticum. PLoS Genet. 13, e1006619 (2017).
Uhlén, M. et al. Proteomics. Tissue-based map of the human proteome. Science 347, 1260419 (2015).
Snyder, E. M. et al. APOBEC1 complementation factor (A1CF) is dispensable for C-to-U RNA editing in vivo. RNA 23, 457–465 (2017).
Conticello, S. G., Thomas, C. J. F., Petersen-Mahrt, S. K. & Neuberger, M. S. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol. Biol. Evol. 22, 367–377 (2005).
Conticello, S. G. The AID/APOBEC family of nucleic acid mutators. Genome Biol. 9, 229 (2008).
Krishnan, A., Iyer, L. M., Holland, S. J., Boehm, T. & Aravind, L. Diversification of AID/APOBEC-like deaminases in metazoa: multiplicity of clades and widespread roles in immunity. Proc. Natl Acad. Sci. USA 115, E3201–E3210 (2018).
Münk, C., Willemsen, A. & Bravo, I. G. An ancient history of gene duplications, fusions and losses in the evolution of APOBEC3 mutators in mammals. BMC Evolut. Biol. 12, 71 (2012).
Hirano, K., Min, J., Funahashi, T., Baunoch, D. A. & Davidson, N. O. Characterization of the human apobec-1 gene: expression in gastrointestinal tissues determined by alternative splicing with production of a novel truncated peptide. J. Lipid Res. 38, 847–859 (1997).
Blanc, V. et al. Genome-wide identification and functional analysis of Apobec-1-mediated C-to-U RNA editing in mouse small intestine and liver. Genome Biol. 15, R79 (2014).
Rosenberg, B. R., Hamilton, C. E., Mwangi, M. M., Dewell, S. & Papavasiliou, F. N. Transcriptome-wide sequencing reveals numerous APOBEC1 mRNA-editing targets in transcript 3′ UTRs. Nat. Struct. Mol. Biol. 18, 230–236 (2011).
Cole, D. C. et al. Loss of APOBEC1 RNA-editing function in microglia exacerbates age-related CNS pathophysiology. Proc. Natl Acad. Sci. USA 114, 13272–13277 (2017).
Niavarani, A., Shahrabi Farahani, A., Sharafkhah, M. & Rassoulzadegan, M. Pancancer analysis identifies prognostic high-APOBEC1 expression level implicated in cancer in-frame insertions and deletions. Carcinogenesis 39, 327–335 (2018).
Rogozin, I. B. et al. Nucleotide weight matrices reveal ubiquitous mutational footprints of AID/APOBEC deaminases in human cancer genomes. Cancers 11, E211 (2019).
Nabel, C. S. et al. AID/APOBEC deaminases disfavor modified cytosines implicated in DNA demethylation. Nat. Chem. Biol. 8, 751–758 (2012).
Rogozin, I. B. & Diaz, M. Cutting edge: DGYW/WRCH is a better predictor of mutability at G:C bases in Ig hypermutation than the widely accepted RGYW/WRCY motif and probably reflects a two-step activation-induced cytidine deaminase-triggered process. J. Immunol. 172, 3382–3384 (2004).
Rogozin, I. B. & Kolchanov, N. A. Somatic hypermutagenesis in immunoglobulin genes. II. Influence of neighbouring base sequences on mutagenesis. Biochim. Biophys. Acta 1171, 11–18 (1992).
Muramatsu, M. et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell 102, 553–563 (2000).
Bransteitter, R., Pham, P., Scharff, M. D. & Goodman, M. F. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc. Natl Acad. Sci. USA 100, 4102–4107 (2003).
Larijani, M. & Martin, A. Single-stranded DNA structure and positional context of the target cytidine determine the enzymatic efficiency of AID. Mol. Cell. Biol. 27, 8038–8048 (2007).
Betz, A. G., Rada, C., Pannell, R., Milstein, C. & Neuberger, M. S. Passenger transgenes reveal intrinsic specificity of the antibody hypermutation mechanism: clustering, polarity, and specific hot spots. Proc. Natl Acad. Sci. USA 90, 2385–2388 (1993).
Rajewsky, K., Forster, I. & Cumano, A. Evolutionary and somatic selection of the antibody repertoire in the mouse. Science 238, 1088–1094 (1987).
Liao, W. et al. APOBEC-2, a cardiac- and skeletal muscle-specific member of the cytidine deaminase supergene family. Biochem. Biophys. Res. Commun. 260, 398–404 (1999).
Etard, C., Roostalu, U. & Strähle, U. Lack of Apobec2-related proteins causes a dystrophic muscle phenotype in zebrafish embryos. J. Cell Biol. 189, 527–539 (2010).
Sato, Y. et al. Deficiency in APOBEC2 leads to a shift in muscle fiber type, diminished body mass, and myopathy. J. Biol. Chem. 285, 7111–7118 (2010).
Sawyer, S. L., Emerman, M. & Malik, H. S. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2, E275 (2004).
LaRue, R. S. et al. Guidelines for naming nonprimate APOBEC3 genes and proteins. J. Virol. 83, 494–497 (2009).
Bogerd, H. P., Wiegand, H. L., Doehle, B. P., Lueders, K. K. & Cullen, B. R. APOBEC3A and APOBEC3B are potent inhibitors of LTR-retrotransposon function in human cells. Nucleic Acids Res. 34, 89–95 (2006).
Refsland, E. W. & Harris, R. S. The APOBEC3 family of retroelement restriction factors. Curr. Top. Microbiol. Immunol. 371, 1–27 (2013).
Yang, B., Chen, K., Zhang, C., Huang, S. & Zhang, H. Virion-associated uracil DNA glycosylase-2 and apurinic/apyrimidinic endonuclease are involved in the degradation of APOBEC3G-edited nascent HIV-1 DNA. J. Biol. Chem. 282, 11667–11675 (2007).
Rogozin, I. B., Basu, M. K., Jordan, I. K., Pavlov, Y. I. & Koonin, E. V. APOBEC4, a new member of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases predicted by computational analysis. Cell Cycle 4, 1281–1285 (2005).
Marino, D. et al. APOBEC4 enhances the replication of HIV-1. PLoS ONE 11, e0155422 (2016).
Shi, M. et al. Characterization and functional analysis of chicken APOBEC4. Dev. Comp. Immunol. 106, 103631 (2020).
Acknowledgements
Work on RNA editing and modification in the Papavasiliou laboratory is funded by the European Research Council (ERC) (#649019) and the German Research Foundation (DFG) (TRR319-RMaP and SPP1784). The authors thank all members, past and present, of the Papavasiliou laboratory for discussions, and sincerely apologize to the many colleagues whose work could not be cited for reasons of space.
Author information
Authors and Affiliations
Contributions
All authors researched data for the article, made substantial contributions to discussions of the content, and wrote the article. F.N.P. and R.P. edited the manuscript and drafted the final submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Genetics thanks M. Larijani and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Human Protein Atlas: http://www.proteinatlas.org
Glossary
- Base modifications
-
Chemically altered nucleotides within mature RNA molecules.
- dCasRx
-
Catalytically dead RNA-guided RNA-targeting CasRx from Ruminococcus flavefaciens XPD3002. CasRx is another member of the CRISPR family (class 2, VI-D).
- dPspCas13b
-
Catalytically dead RNA-guided RNA-targeting CRISPR–Cas13b from Prevotella sp. P5-125. Cas13b is a member of the CRISPR family (class 2, type VI). Physiologically, Cas13b catalyses site-specific cleavage of single-stranded RNA.
- G-quadruplex
-
A non-canonical four-stranded secondary structure of guanine-rich DNA sequences.
- Guide RNAs
-
(gRNAs). Short RNA sequences used in base-editing technologies to target the base editor to a specific sequence in DNA or RNA. Depending on the tagging system used, the base editor can be recruited by the gRNA using specific scaffolds (for Cas proteins), sequences (MS2 coat protein) or chemical modifications (for SNAP).
- Intrinsically disordered region
-
(IDR). An unstructured domain of proteins that are believed to have roles in intermolecular and intramolecular interactions, such as complex formation and phase separation.
- MS2-tagged
-
Refers to a molecule labelled using a tagging system based on the natural interaction between the MS2 bacteriophage coat protein and a stem–loop structure from the phage genome. The sequence forming the stem–loop can be attached to a guide RNA (gRNA) to target an MS2-tagged base editor.
- Nuclear export signal
-
(NES). A short peptide motif enriched for hydrophobic residues (such as Leu) recognized by exportins (such as XPO1/CRM1) that tags a protein for nuclear exit.
- Nuclear localization signal
-
(NLS). A short peptide motif enriched for positively charged residues that tags a protein for nuclear import.
- Pseudotyped HIV
-
Chimaeric viruses composed of the envelope glycoprotein of vesicular stomatis virus (VSV-G) and the human immunodeficiency virus type 1 (HIV-1) core; these viruses are more infectious than non-pseudotyped HIV-1 viruses.
- SNAP-tagged
-
Refers to a molecule labelled using a tagging system based on the SNAP-tag self-labelling protein derived from the human O6-alkylguanine-DNA alkyltransferase. As a SNAP-tag will form a covalent linkage with benzylguanine (BG)-modified nucleotides, a SNAP-tagged base editor can be directed to specific targets by BG-modified guide RNAs (gRNAs).
- Stem–loops
-
Specific structures that may occur in single-stranded RNA (ssRNA) when complementary sequences base pair to form a double helix that ends in an unpaired (single-stranded) loop. Stem–loops are also known as hairpin structures or hairpin loops.
- π-Stacking
-
Attractive non-covalent interactions between aromatic rings.
- Tumour restriction
-
The limitation of tumour growth and/or tumour suppression or ablation by numerous distinct molecular mechanisms. Here, we specifically refer to the limitation of tumour growth owing to cell death after activation-induced cytidine deaminase/apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like (AID/APOBEC)-mediated hypermutation.
Rights and permissions
About this article
Cite this article
Pecori, R., Di Giorgio, S., Paulo Lorenzo, J. et al. Functions and consequences of AID/APOBEC-mediated DNA and RNA deamination. Nat Rev Genet 23, 505–518 (2022). https://doi.org/10.1038/s41576-022-00459-8
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41576-022-00459-8
This article is cited by
-
A proteogenomic atlas of 1032 brain metastases identifies molecular subtypes, immune landscapes, and therapeutic vulnerabilities
Nature Communications (2026)
-
DOT1L activity limits transcription elongation velocity and favors RNAPII pausing to facilitate mutagenesis by AID
Nature Communications (2026)
-
The expanding roles of homologous recombination proteins in genome stability
The EMBO Journal (2026)
-
Single-strand deaminase-assisted editing for functional RNA manipulation
Nature Biotechnology (2026)
-
Action-at-a-distance mutations by 8-oxo-7,8-dihydroguanine: adenine pair triggered by MUTYH
Genes and Environment (2025)