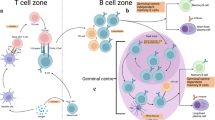
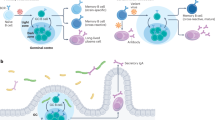
Abstract
Memory B cells (MBCs) are critical for the rapid development of protective immunity following re-infection. MBCs capable of neutralizing distinct subclasses of pathogens, such as influenza and HIV, have been identified in humans. However, efforts to develop vaccines that induce broadly protective MBCs to rapidly mutating pathogens have not yet been successful. Better understanding of the signals regulating MBC development and function are essential to overcome current challenges hindering successful vaccine development. Here, we discuss recent advancements regarding the signals and transcription factors regulating germinal centre-derived MBC development and function.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Victora, G. D. et al. Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell 143, 592–605 (2010).
Dogan, I. et al. Multiple layers of B cell memory with different effector functions. Nat. Immunol. 10, 1292–1299 (2009).
McHeyzer-Williams, L. J., Milpied, P. J., Okitsu, S. L. & McHeyzer-Williams, M. G. Class-switched memory B cells remodel BCRs within secondary germinal centers. Nat. Immunol. 16, 296–305 (2015).
Pape, K. A., Taylor, J. J., Maul, R. W., Gearhart, P. J. & Jenkins, M. K. Different B cell populations mediate early and late memory during an endogenous immune response. Science 331, 1203–1207 (2011).
Zuccarino-Catania, G. V. et al. CD80 and PD-L2 define functionally distinct memory B cell subsets that are independent of antibody isotype. Nat. Immunol. 15, 631–637 (2014). This study identifies new markers that could identify transcriptionally distinct MBC subsets with differing capacities to differentiate into antibody-secreting cells or GC B cells upon antigen re-encounter.
Weisel, F. J., Zuccarino-Catania, G. V., Chikina, M. & Shlomchik, M. J. A temporal switch in the germinal center determines differential output of memory B and plasma cells. Immunity 44, 116–130 (2016). This paper uses a pulse-labelling approach to show that MBC export from the GC precedes that of plasma cells.
Shinnakasu, R. et al. Regulated selection of germinal-center cells into the memory B cell compartment. Nat. Immunol. 17, 861–869 (2016). This study proposes an instructive model of MBC development in which weak T cell help predisposes GC B cells to express BACH2 and enter the memory compartment.
Laidlaw, B. J., Duan, L., Xu, Y., Vazquez, S. E. & Cyster, J. G. The transcription factor Hhex cooperates with the corepressor Tle3 to promote memory B cell development. Nat. Immunol 21, 1082–1093 (2020). Using CRISPR–Cas9 screening in vivo, this study found that HHEX and TLE3 cooperate to promote MBC development during a viral response.
Purtha, W. E., Tedder, T. F., Johnson, S., Bhattacharya, D. & Diamond, M. S. Memory B cells, but not long-lived plasma cells, possess antigen specificities for viral escape mutants. J. Exp. Med. 208, 2599–2606 (2011). This study finds that MBCs, but not plasma cells, can recognize previously unencountered stains of the West Nile virus, suggesting that MBCs might be an important component of heterosubtypic immunity.
Andrade, P. et al. Impact of pre-existing dengue immunity on human antibody and memory B cell responses to Zika. Nat. Commun. 10, 938 (2019).
McCarthy, K. R. et al. Memory B cells that cross-react with group 1 and group 2 influenza A viruses are abundant in adult human repertoires. Immunity 48, 174–183.e9 (2018).
Leach, S. et al. Requirement for memory B-cell activation in protection from heterologous influenza virus reinfection. Int. Immunol. 31, 771–779 (2019).
Benson, M. J. et al. Distinction of the memory B cell response to cognate antigen versus bystander inflammatory signals. J. Exp. Med. 206, 2013–2025 (2009).
Krishnamurty, A. T. et al. Somatically hypermutated plasmodium-specific IgM+ memory B cells are rapid, plastic, early responders upon malaria rechallenge. Immunity 45, 402–414 (2016).
Zabel, F. et al. Viral particles drive rapid differentiation of memory B cells into secondary plasma cells producing increased levels of antibodies. J. Immunol. 192, 5499–5508 (2014).
Mesin, L. et al. Restricted clonality and limited germinal center reentry characterize memory B cell reactivation by boosting. Cell 180, 92–106.e11 (2020). This study finds that most MBC clones fail to re-enter the GC during recall responses. Rather, recall GCs tend to consist largely of B cells with no GC experience.
Zhang, Y. et al. Germinal center B cells govern their own fate via antibody feedback. J. Exp. Med. 210, 457–464 (2013).
Toyama, H. et al. Memory B cells without somatic hypermutation are generated from Bcl6-deficient B cells. Immunity 17, 329–339 (2002).
Obukhanych, T. V. & Nussenzweig, M. C. T-independent type II immune responses generate memory B cells. J. Exp. Med. 203, 305–310 (2006).
Pritchard, G. H. et al. The development of optimally responsive plasmodium-specific CD73+CD80+IgM+ memory B cells requires intrinsic BCL6 expression but not CD4 TFH cells. Preprint at bioRxiv https://doi.org/10.1101/564351 (2019).
Kenderes, K. J. et al. T-Bet+ IgM memory cells generate multi-lineage effector B cells. Cell Rep. 24, 824–837.e3 (2018).
Fukuda, T. et al. Disruption of the Bcl6 gene results in an impaired germinal center formation. J. Exp. Med. 186, 439–448 (1997).
Huang, C. et al. The BCL6 RD2 domain governs commitment of activated B cells to form germinal centers. Cell Rep. 8, 1497–1508 (2014).
Willis, S. N. et al. Bcl6 protein expression shapes pre-germinal center B cell dynamics and follicular helper T cell heterogeneity. J. Immunol. 192, 3200–3206 (2011).
Basso, K. et al. Integrated biochemical and computational approach identifies BCL6 direct target genes controlling multiple pathways in normal germinal center B cells. Blood 115, 975–984 (2010).
Saito, M. et al. BCL6 suppression of BCL2 via Miz1 and its disruption in diffuse large B cell lymphoma. Proc. Natl Acad. Sci. USA 106, 11294–11299 (2009).
Li, Z. et al. BCL-6 negatively regulates expression of the NF-κB1 p105/p50 subunit. J. Immunol. 174, 205–214 (2004).
Linterman, M. A. et al. IL-21 acts directly on B cells to regulate Bcl-6 expression and germinal center responses. J. Exp. Med. 207, 353–363 (2010).
Chevrier, S., Kratina, T., Emslie, D., Tarlinton, D. M. & Corcoran, L. M. IL4 and IL21 cooperate to induce the high Bcl6 protein level required for germinal center formation. Immunol. Cell Biol. https://doi.org/10.1038/icb.2017.71 (2017).
Zotos, D. et al. IL-21 regulates germinal center B cell differentiation and proliferation through a B cell-intrinsic mechanism. J. Exp. Med. 207, 365–378 (2010).
Avery, D. T. et al. B cell-intrinsic signaling through IL-21 receptor and STAT3 is required for establishing long-lived antibody responses in humans. J. Exp. Med. 207, 155–171 (2010).
Ochiai, K. et al. Transcriptional regulation of germinal center B and plasma cell fates by dynamical control of IRF4. Immunity 38, 918–929 (2013).
Klein, U. et al. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat. Immunol. 7, 773–782 (2006).
Willis, S. N. et al. Transcription factor IRF4 regulates germinal center cell formation through a B cell-intrinsic mechanism. J. Immunol. 192, 3200–3206 (2014).
Sciammas, R. et al. An incoherent regulatory network architecture that orchestrates B cell diversification in response to antigen signaling. Mol. Syst. Biol. 7, 495 (2011).
Saito, M. et al. A signaling pathway mediating downregulation of BCL6 in germinal center B cells is blocked by BCL6 gene alterations in B cell lymphoma. Cancer Cell 12, 280–292 (2007).
Sciammas, R. et al. Graded expression of interferon regulatory factor-4 coordinates isotype switching with plasma cell differentiation. Immunity 25, 225–236 (2006).
Lee, C. H. et al. Regulation of the germinal center gene program by interferon (IFN) regulatory factor 8/IFN consensus sequence-binding protein. J. Exp. Med. 203, 63–72 (2006).
Zhou, J. X. et al. IFN regulatory factor 8 regulates MDM2 in germinal center B cells. J. Immunol. 183, 3188–3194 (2009).
Yoon, J. et al. Interferon regulatory factor 8 (IRF8) interacts with the B cell lymphoma 6 (BCL6) corepressor BCOR. J. Biol. Chem. 289, 34250–34257 (2014).
Carotta, S. et al. The transcription factors IRF8 and PU.1 negatively regulate plasma cell differentiation. J. Exp. Med. 211, 2169–2181 (2014).
Wang, H. et al. Transcription factors IRF8 and PU.1 are required for follicular B cell development and BCL6-driven germinal center responses. Proc. Natl Acad. Sci. USA 116, 9511–9520 (2019).
Schubart, D. B., Rolink, A., Kosco-Vilbois, M. H., Botteri, F. & Matthias, P. B-cell-specific coactivator OBF-1/OCA-B/Bob1 required for immune response and germinal centre formation. Nature 383, 538–542 (1996).
Kim, U. et al. The B-cell-specific transcription coactivator OCA-B/OBF-1/Bob-1 is essential for normal production of immunoglobulin isotypes. Nature 383, 542–547 (1996).
Corcoran, L. et al. Oct2 and Obf1 as facilitators of B:T cell collaboration during a humoral immune response. Front. Immunol. 5, 108 (2014).
Karnowski, A. et al. B and T cells collaborate in antiviral responses via IL-6, IL-21, and transcriptional activator and coactivator, Oct2 and OBF-1. J. Exp. Med. 209, 2049–2064 (2012).
Bartholdy, B. et al. The Ets factor Spi-B is a direct critical target of the coactivator OBF-1. Proc. Natl Acad. Sci. USA 103, 11665–11670 (2006).
Garrett-Sinha, L. A. et al. PU.1 and Spi-B are required for normal B cell receptor-mediated signal transduction. Immunity 10, 399–408 (1999).
Su, G. H. et al. Defective B cell receptor-mediated responses in mice lacking the Ets protein, Spi-B. EMBO J. 16, 7118–7129 (1997).
Willis, S. N. et al. Environmental sensing by mature B cells is controlled by the transcription factors PU.1 and SpiB. Nat. Commun. 8, 1426 (2017).
Calado, D. P. et al. The cell-cycle regulator c-Myc is essential for the formation and maintenance of germinal centers. Nat. Immunol. 13, 1092–1100 (2012).
Dominguez-Sola, D. et al. The proto-oncogene MYC is required for selection in the germinal center and cyclic reentry. Nat. Immunol. 13, 1083–1091 (2012).
Tanaka, H. et al. E2F1 and c-Myc potentiate apoptosis through inhibition of NF-κB activity that facilitates MnSOD-mediated ROS elimination. Mol. Cell 9, 1017–1029 (2002).
Béguelin, W. et al. EZH2 enables germinal centre formation through epigenetic silencing of CDKN1A and an Rb–E2F1 feedback loop. Nat. Commun. 8, 877 (2017).
Brescia, P. et al. MEF2B instructs germinal center development and acts as an oncogene in B cell lymphomagenesis. Cancer Cell 34, 453–465.e9 (2018).
Wilker, P. R. et al. Transcription factor Mef2c is required for B cell proliferation and survival after antigen receptor stimulation. Nat. Immunol. 9, 603–612 (2008).
Khiem, D., Cyster, J. G., Schwarz, J. J. & Black, B. L. A p38 MAPK–MEF2C pathway regulates B-cell proliferation. Proc. Natl Acad. Sci. USA 105, 17067–17072 (2008).
Ying, C. Y. et al. MEF2B mutations lead to deregulated expression of the oncogene BCL6 in diffuse large B cell lymphoma. Nat. Immunol. 14, 1084–1092 (2013).
Kwon, K. et al. Instructive role of the transcription factor E2A in early B lymphopoiesis and germinal center B cell development. Immunity 28, 751–762 (2008).
Wöhner, M. et al. Molecular functions of the transcription factors E2A and E2-2 in controlling germinal center B cell and plasma cell development. J. Exp. Med. 213, 1201–1221 (2016).
Gloury, R. et al. Dynamic changes in Id3 and E-protein activity orchestrate germinal center and plasma cell development. J. Exp. Med. 213, 1095–1111 (2016).
Chen, S. et al. Id3 orchestrates germinal center B cell development. Mol. Cell. Biol. 36, 2543–2552 (2016).
Green, M. R. et al. Signatures of murine B-cell development implicate Yy1 as a regulator of the germinal center-specific program. Proc. Natl Acad. Sci. USA 108, 2873–2878 (2011).
Vilagos, B. et al. Essential role of EBF1 in the generation and function of distinct mature B cell types. J. Exp. Med. 209, 775–792 (2012).
Gyory, I. et al. Transcription factor Ebf1 regulates differentiation stage-specific signaling, proliferation, and survival of B cells. Genes. Dev. 26, 668–682 (2012).
Huang, C., Geng, H., Boss, I., Wang, L. & Melnick, A. Cooperative transcriptional repression by BCL6 and BACH2 in germinal center B-cell differentiation. Blood 123, 1012–1020 (2014).
Shaffer, A. L. et al. BCL-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity 13, 199–212 (2000).
Reljic, R., Wagner, S. D., Peakman, L. J. & Fearon, D. T. Suppression of signal transducer and activator of transcription 3-dependent B lymphocyte terminal differentiation by BCL-6. J. Exp. Med. 192, 1841–1848 (2000).
Polo, J. M., Ci, W., Licht, J. D. & Melnick, A. Reversible disruption of BCL6 repression complexes by CD40 signaling in normal and malignant B cells. Blood 112, 644–651 (2008).
Niu, H., Ye, B. H. & Dalla-Favera, R. Antigen receptor signaling induces MAP kinase-mediated phosphorylation and degradation of the BCL-6 transcription factor. Genes. Dev. 12, 1953–1961 (1998).
Zhang, T.-T. et al. Germinal center B cell development has distinctly regulated stages completed by disengagement from T cell help. eLife 6, e19552 (2017).
Ise, W. et al. T follicular helper cell-germinal center B cell interaction strength regulates entry into plasma cell or recycling germinal center cell fate. Immunity 48, 702–715.e4 (2018).
Weinstein, J. S. et al. TFH cells progressively differentiate to regulate the germinal center response. Nat. Immunol. 17, 1197–1205 (2016).
Li, X. et al. Cbl ubiquitin ligases control B cell exit from the germinal-center reaction. Immunity 48, 530–541.e6 (2018).
Davidzohn, N. et al. Syk degradation restrains plasma cell formation and promotes zonal transitions in germinal centers. J. Exp. Med. 217, 26648 (2019).
Sohn, H. W., Gu, H. & Pierce, S. K. Cbl-b negatively regulates B cell antigen receptor signaling in mature B cells through ubiquitination of the tyrosine kinase Syk. J. Exp. Med. 197, 1511–1524 (2003).
Ly, A. et al. Transcription factor T-bet in B cells modulates germinal center polarization and antibody affinity maturation in response to malaria. Cell Rep. 29, 2257–2269.e6 (2019).
Guthmiller, J. J., Graham, A. C., Zander, R. A., Pope, R. L. & Butler, N. S. Cutting edge: IL-10 is essential for the generation of germinal center B cell responses and anti-plasmodium humoral immunity. J. Immunol. 198, 617–622 (2017).
Rubtsova, K. et al. B cells expressing the transcription factor T-bet drive lupus-like autoimmunity. J. Clin. Invest. 127, 1392–1404 (2017).
Jackson, S. W. et al. B cell IFN-γ receptor signaling promotes autoimmune germinal centers via cell-intrinsic induction of BCL-6. J. Exp. Med. 213, 733–750 (2016).
Sander, S. et al. PI3 kinase and FOXO1 transcription factor activity differentially control B cells in the germinal center light and dark zones. Immunity 43, 1075–1086 (2015).
Dominguez-Sola, D. et al. The FOXO1 transcription factor instructs the germinal center dark zone program. Immunity 43, 1064–1074 (2015).
Inoue, T. et al. The transcription factor Foxo1 controls germinal center B cell proliferation in response to T cell help. J. Exp. Med. 214, 1181–1198 (2017).
Bannard, O. et al. Germinal center centroblasts transition to a centrocyte phenotype according to a timed program and depend on the dark zone for effective selection. Immunity 39, 912–924 (2013).
Albright, A. R. et al. TGFβ signaling in germinal center B cells promotes the transition from light zone to dark zone. J. Exp. Med. 216, 2531–2545 (2019).
Laidlaw, B. J. et al. Interleukin-10 from CD4+ follicular regulatory T cells promotes the germinal center response. Sci. Immunol. 2, eaan4767 (2017).
Koike, T., Harada, K., Horiuchi, S. & Kitamura, D. The quantity of CD40 signaling determines the differentiation of B cells into functionally distinct memory cell subsets. eLife 8, e44245 (2019).
Ise, W. et al. The transcription factor BATF controls the global regulators of class-switch recombination in both B cells and T cells. Nat. Immunol. 12, 536–543 (2011).
Finkin, S., Hartweger, H., Oliveira, T. Y., Kara, E. E. & Nussenzweig, M. C. Protein amounts of the MYC transcription factor determine germinal center B cell division capacity. Immunity 51, 324–336.e5 (2019).
Chen, C. et al. Uhrf1 regulates germinal center B cell expansion and affinity maturation to control viral infection. J. Exp. Med. 215, 1437–1448 (2018).
Toboso-Navasa, A. et al. Restriction of memory B cell differentiation at the germinal center B cell positive selection stage. J. Exp. Med. 217, 943 (2020).
Chou, C. et al. The transcription factor AP4 mediates resolution of chronic viral infection through amplification of germinal center B cell responses. Immunity 45, 570–582 (2016).
Luo, W., Weisel, F. & Shlomchik, M. J. B cell receptor and CD40 signaling are rewired for synergistic induction of the c-Myc transcription factor in germinal center B cells. Immunity 48, 313–326.e5 (2018).
Gallagher, E. et al. Kinase MEKK1 is required for CD40-dependent activation of the kinases Jnk and p38, germinal center formation, B cell proliferation and antibody production. Nat. Immunol. 8, 57–63 (2006).
Heise, N. et al. Germinal center B cell maintenance and differentiation are controlled by distinct NF-κB transcription factor subunits. J. Exp. Med. 211, 2103–2118 (2014).
Grumont, R. J. & Gerondakis, S. Rel induces interferon regulatory factor 4 (IRF-4) expression in lymphocytes: modulation of interferon-regulated gene expression by rel/nuclear factor-κB. J. Exp. Med. 191, 1281–1292 (2000).
Grumont, R. J., Strasser, A. & Gerondakis, S. B cell growth is controlled by phosphatidylinosotol 3-kinase-dependent induction of Rel/NF-κB regulated c-myc transcription. Mol. Cell 10, 1283–1294 (2002).
De Silva, N. S. et al. Transcription factors of the alternative NF-κB pathway are required for germinal center B-cell development. Proc. Natl Acad. Sci. USA 113, 9063–9068 (2016).
Luo, W. et al. The AKT kinase signaling network is rewired by PTEN to control proximal BCR signaling in germinal center B cells. Nat. Immunol. 20, 736–746 (2019).
Zhu, Z., Shukla, A., Ramezani-Rad, P., Apgar, J. R. & Rickert, R. C. The AKT isoforms 1 and 2 drive B cell fate decisions during the germinal center response. Life Sci. Alliance 2, e201900506 (2019).
Ersching, J. et al. Germinal center selection and affinity maturation require dynamic regulation of mTORC1 kinase. Immunity 46, 1045–1058.e6 (2017).
Raybuck, A. L. et al. B cell-intrinsic mTORC1 promotes germinal center-defining transcription factor gene expression, somatic hypermutation, and memory B cell generation in humoral immunity. J. Immunol. 200, 2627–2639 (2018).
Kometani, K. et al. Repression of the transcription factor Bach2 contributes to predisposition of IgG1 memory B cells toward plasma cell differentiation. Immunity 39, 136–147 (2013).
Mayer, C. T. et al. The microanatomic segregation of selection by apoptosis in the germinal center. Science 358, eaao2602 (2017).
Stewart, I., Radtke, D., Phillips, B., McGowan, S. J. & Bannard, O. Germinal center B cells replace their antigen receptors in dark zones and fail light zone entry when immunoglobulin gene mutations are damaging. Immunity 49, 477–489.e7 (2018).
Tamahara, T. et al. The mTOR–Bach2 cascade controls cell cycle and class switch recombination during B cell differentiation. Mol. Cell. Biol. 37, e00418-17 (2017).
Miura, Y. et al. Bach2 promotes B cell receptor-induced proliferation of B lymphocytes and represses cyclin-dependent kinase inhibitors. J. Immunol. 200, 2882–2893 (2018).
Fischer, S. F. et al. Proapoptotic BH3-only protein Bim is essential for developmentally programmed death of germinal center-derived memory B cells and antibody-forming cells. Blood 110, 3978–3984 (2007).
Smith, K. G., Weiss, U., Rajewsky, K., Nossal, G. J. & Tarlinton, D. M. Bcl-2 increases memory B cell recruitment but does not perturb selection in germinal centers. Immunity 1, 803–813 (1994).
Laidlaw, B. J. et al. The Eph-related tyrosine kinase ligand Ephrin-B1 marks germinal center and memory precursor B cells. J. Exp. Med. 214, 639–649 (2017).
Wang, Y. et al. Germinal-center development of memory B cells driven by IL-9 from follicular helper T cells. Nat. Immunol. 18, 921–930 (2017).
Suan, D. et al. CCR6 defines memory B cell precursors in mouse and human germinal centers, revealing light-zone location and predominant low antigen affinity. Immunity 47, 1142–1153.e4 (2017). Laidlaw et al. (J. Exp. Med., 2017), Wang et al. (2017) and Suan et al. identify and characterize MBC precursors residing in the light zone of the GC in mice and humans.
Kuo, T. C. et al. Repression of BCL-6 is required for the formation of human memory B cells in vitro. J. Exp. Med. 204, 819–830 (2007).
Hatzi, K. et al. A hybrid mechanism of action for BCL6 in B cells defined by formation of functionally distinct complexes at enhancers and promoters. Cell Rep. 4, 578–588 (2013).
Bhattacharya, D. et al. Transcriptional profiling of antigen-dependent murine B cell differentiation and memory formation. J. Immunol. 179, 6808–6819 (2007).
Xing, S. et al. Tle corepressors are differentially partitioned to instruct CD8+ T cell lineage choice and identity. J. Exp. Med. 215, 2211–2226 (2018).
Jangal, M. et al. The transcriptional co-repressor TLE3 suppresses basal signaling on a subset of estrogen receptor α target genes. Nucleic Acids Res. 42, 11339–11348 (2014).
Takatsuka, S. et al. IL-9 receptor signaling in memory B cells regulates humoral recall responses. Nat. Immunol. 19, 1025–1034 (2018).
Jash, A. et al. ZBTB32 restricts the duration of memory B cell recall responses. J. Immunol. 197, 1159–1168 (2016).
Bernasconi, N. L., Onai, N. & Lanzavecchia, A. A role for Toll-like receptors in acquired immunity: up-regulation of TLR9 by BCR triggering in naive B cells and constitutive expression in memory B cells. Blood 101, 4500–4504 (2003).
Onodera, T. Whole-virion influenza vaccine recalls an early burst of high-affinity memory B cell response through TLR signaling. J. Immunol. 196, 4172–4184 (2016).
Lutz, J. et al. Reactivation of IgG-switched memory B cells by BCR-intrinsic signal amplification promotes IgG antibody production. Nat. Commun. 6, 8575 (2015).
Thomas, M. J., Klein, U., Lygeros, J. & Rodríguez Martínez, M. A probabilistic model of the germinal center reaction. Front. Immunol. 10, 689 (2019).
Wang, P., Shih, C.-M., Qi, H. & Lan, Y.-H. A stochastic model of the germinal center integrating local antigen competition, individualistic T–B interactions, and B cell receptor signaling. J. Immunol. 197, 1169–1182 (2016).
Vikstrom, I. et al. Mcl-1 is essential for germinal center formation and B cell memory. Science 330, 1095–1099 (2010).
Barnett, B. E. et al. Asymmetric B cell division in the germinal center reaction. Science 335, 342–344 (2012).
Lin, W.-H. W. et al. Asymmetric PI3K signaling driving developmental and regenerative cell fate bifurcation. Cell Rep. https://doi.org/10.1016/j.celrep.2015.10.072 (2015).
Duffy, K. R. et al. Activation-induced B cell fates are selected by intracellular stochastic competition. Science 335, 338–341 (2012).
Taylor, J. J., Pape, K. A., Steach, H. R. & Jenkins, M. K. Humoral immunity. Apoptosis and antigen affinity limit effector cell differentiation of a single naïve B cell. Science 347, 784–787 (2015).
Tas, J. M. J. et al. Visualizing antibody affinity maturation in germinal centers. Science 351, 1048–1054 (2016).
Allie, S. R. et al. The establishment of resident memory B cells in the lung requires local antigen encounter. Nat. Immunol. 20, 97–108 (2019). This study finds that MBCs develop in the lungs after influenza infection and rapidly differentiate into local antibody-secreting cells upon challenge infection.
Adachi, Y. et al. Distinct germinal center selection at local sites shapes memory B cell response to viral escape. J. Exp. Med. 212, 1709–1723 (2015). This study finds that MBCs in the lungs have an elevated fraction of cross-reactive clones relative to cells in the secondary lymphoid organs after influenza virus infection.
Sungalee, S. et al. Germinal center reentries of BCL2-overexpressing B cells drive follicular lymphoma progression. J. Clin. Invest. 124, 5337–5351 (2014).
Schmitz, R. et al. Genetics and pathogenesis of diffuse large B-cell lymphoma. N. Engl. J. Med. 378, 1396–1407 (2018).
Chapuy, B. et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat. Med. 24, 679–690 (2018).
Venturutti, L. et al. TBL1XR1 mutations drive extranodal lymphoma by inducing a pro-tumorigenic memory fate. Cell https://doi.org/10.1016/j.cell.2020.05.049 (2020). This paper reports that a subset of B cell lymphoma is driven by mutations that bias cells to develop into immature MBCs.
He, J.-S. et al. IgG1 memory B cells keep the memory of IgE responses. Nat. Commun. 8, 641 (2017).
Asrat, S. et al. Chronic allergen exposure drives accumulation of long-lived IgE plasma cells in the bone marrow, giving rise to serological memory. Sci. Immunol. 5, eaav8402 (2020).
Gowthaman, U. et al. Identification of a T follicular helper cell subset that drives anaphylactic IgE. Science 365, eaaw6433 (2019).
Sayegh, C. E., Quong, M. W., Agata, Y. & Murre, C. E-proteins directly regulate expression of activation-induced deaminase in mature B cells. Nat. Immunol. 4, 586–593 (2003).
Gonda, H. et al. The balance between Pax5 and Id2 activities is the key to AID gene expression. J. Exp. Med. 198, 1427–1437 (2003).
Tomayko, M. M., Steinel, N. C., Anderson, S. M. & Shlomchik, M. J. Cutting edge: hierarchy of maturity of murine memory B cell subsets. J. Immunol. 185, 7146–7150 (2010).
Onodera, T. et al. Memory B cells in the lung participate in protective humoral immune responses to pulmonary influenza virus reinfection. Proc. Natl Acad. Sci. USA 109, 2485–2490 (2012).
Lindner, C. et al. Diversification of memory B cells drives the continuous adaptation of secretory antibodies to gut microbiota. Nat. Immunol. 16, 880–888 (2015).
Bar-Or, A. et al. Immunological memory: contribution of memory B cells expressing costimulatory molecules in the resting state. J. Immunol. 167, 5669–5677 (2001).
Weisel, F. & Shlomchik, M. Memory B cells of mice and humans. Annu. Rev. Immunol. 35, 255–284 (2017).
Seifert, M. et al. Functional capacities of human IgM memory B cells in early inflammatory responses and secondary germinal center reactions. Proc. Natl Acad. Sci. USA 112, E546–E555 (2015).
Siewe, B., Nipper, A. J., Sohn, H., Stapleton, J. T. & Landay, A. FcRL4 expression identifies a pro-inflammatory B cell subset in viremic HIV-infected subjects. Front. Immunol. 8, 117 (2017).
Naradikian, M. S. et al. Cutting edge: IL-4, IL-21, and IFN-γ interact to govern T-bet and CD11c expression in TLR-activated B cells. J. Immunol. 197, 1023–1028 (2016).
Yates, J. L., Racine, R., McBride, K. M. & Winslow, G. M. T Cell–Dependent IgM Memory B Cells generated during bacterial infection are required for IgG responses to antigen challenge. J. Immunol. 191, 1240–1249 (2013).
Jenks, S. A. et al. Distinct effector B cells induced by unregulated Toll-like receptor 7 contribute to pathogenic responses in systemic lupus erythematosus. Immunity 49, 725–739.e6 (2018).
Du, S. W. et al. Functional characterization of CD11c+ age-associated B cells as memory B cells. J. Immunol. 203, 2817–2826 (2019).
Kim, C. C., Baccarella, A. M., Bayat, A., Pepper, M. & Fontana, M. F. FCRL5+ memory B cells exhibit robust recall responses. Cell Rep. 27, 1446–1460.e4 (2019).
Peng, S. L., Szabo, S. J. & Glimcher, L. H. T-bet regulates IgG class switching and pathogenic autoantibody production. Proc. Natl Acad. Sci. USA 99, 5545–5550 (2002).
Johnson, J. L. et al. The transcription factor T-bet resolves memory B cell subsets with distinct tissue distributions and antibody specificities in mice and humans. Immunity 52, 841–855.e6 (2020).
Piovesan, D. et al. c-Myb regulates the T-bet-dependent differentiation program in B cells to coordinate antibody responses. Cell Rep. 19, 461–470 (2017).
Stone, S. L. et al. T-bet transcription factor promotes antibody-secreting cell differentiation by limiting the inflammatory effects of IFN-γ on B cells. Immunity 50, 1172–1187.e7 (2019).
Rubtsova, K., Rubtsov, A. V., van Dyk, L. F., Kappler, J. W. & Marrack, P. T-box transcription factor T-bet, a key player in a unique type of B-cell activation essential for effective viral clearance. Proc. Natl Acad. Sci. USA 110, E3216–E3224 (2013).
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information
Nature Reviews Immunology thank the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Germinal centre
-
A structure that forms in the B cell follicle of secondary lymphoid organs during an immune response in which B cells engage with antigen and compete for signals necessary for their survival, proliferation and differentiation into plasma cells or memory B cells.
- Somatic hypermutation
-
A cellular process in which proliferating B cells accumulate mutations in antibody complementarity-determining regions, potentially impacting their ability to recognize antigen.
- Plasma cells
-
Large, terminally differentiated B lymphocytes that continually secrete antibodies and are also known as antibody-secreting cells.
- Recall responses
-
Immune responses in which memory B cells re-encounter their cognate antigen and differentiate into antibody-secreting cells or re-enter the germinal centre to undergo further diversification.
- Affinity maturation
-
The process by which germinal centre B cells increase their affinity for antigen during an immune response as a result of competition for a limiting amount of CD40L-expressing follicular helper T cells.
- Cross-reactive viral antigens
-
Viral protein sequences that are conserved between different strains of a pathogen.
- Heterosubtypic immunity
-
Immunity in which lymphocytes are generated that can protect against multiple subtypes of a pathogen.
- MBC clones
-
Memory B cells (MBCs) with a particular specificity for antigen.
- Secondary GC response
-
A new germinal centre (GC) that forms against an immunogen or pathogen that the host previously encountered and developed immunological memory against.
- Naive B cells
-
B cells that have not been exposed to antigen.
- Class-switched
-
A cellular process in which proliferating B cells have rearranged their constant region genes to switch from expressing one class of immunoglobulin to another, without altering their antigen specificity.
- Dark zone
-
The compartment of the germinal centre in which B cells proliferate and undergo somatic hypermutation, which contains a network of stromal cells producing the CXCR4 ligand CXCL12.
- Light zone
-
The compartment of the germinal centre in which B cells capture antigen presented by follicular dendritic cells and compete to present antigen to follicular helper T cells in order to receive signals necessary for their continued survival, proliferation and differentiation.
- Type 2 immune responses
-
An immune response that typically occurs in response to extracellular bacteria, parasites, toxins or allergens. Generally characterized by the production of IL-4, IL-5 and IL-13.
Rights and permissions
About this article
Cite this article
Laidlaw, B.J., Cyster, J.G. Transcriptional regulation of memory B cell differentiation. Nat Rev Immunol 21, 209–220 (2021). https://doi.org/10.1038/s41577-020-00446-2
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41577-020-00446-2
This article is cited by
-
Integration of functional genomics and statistical fine-mapping systematically characterizes adult-onset and childhood-onset asthma genetic associations
Genome Medicine (2025)
-
B cells in non-lymphoid tissues
Nature Reviews Immunology (2025)
-
NF-κB inhibition attenuates sympathetic hyperreflexia and concomitant development of autonomic dysreflexia and immune dysfunction after spinal cord injury
Communications Biology (2025)
-
The role of fatty acid metabolism on B cells and B cell-related autoimmune diseases
Inflammation Research (2025)
-
Molecular Mechanisms Behind Organ-Related Aging and Regulatory Effects of Natural Therapeutics
Cell Biochemistry and Biophysics (2025)