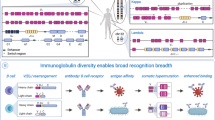
Abstract
The mystery surrounding the mechanisms by which antibody diversity is generated was largely settled in the 1970s by the discoveries of variable gene rearrangements and somatic hypermutation. This led to the paradigm that immunoglobulins are composed of two independent domains — variable and constant — that confer specificity and effector functions, respectively. However, since these early discoveries, there have been a series of observations of communication between the variable and constant domains that affects the overall antibody structure, which suggests that immunoglobulins have a more complex, interconnected functionality than previously thought. Another unresolved issue has been the genesis of ‘restricted’ antibody responses, characterized by the use of only a few variable region gene segments, despite the enormous potential combinatorial diversity. In this Perspective, we place recent findings related to immunoglobulin structure and function in the context of these immunologically important, historically unsolved problems to propose a new model for how antibody specificity is achieved without autoreactivity.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Silverstein, A. M. Splitting the difference: the germline-somatic mutation debate on generating antibody diversity. Nat. Immunol. 4, 829–833 (2003).
Sigal, N. H. & Klinman, N. R. The B-cell clonotype repertoire. Adv. Immunol. 26, 255–337 (1978).
Kreth, H. W. & Williamson, A. R. The extent of diversity of anti-hapten antibodies in inbred mice: anti-NIP (4-hydroxy-5-iodo-3-nitro-phenacetyl) antibodies in CBA/H mice. Eur. J. Immunol. 3, 141–146 (1973).
Pink, J. R. L. & Askonas, B. A. Diversity of antibodies to cross-reacting nitrophenyl haptens in inbred mice. Eur. J. Immunol. 4, 426–430 (1974).
Yung, L. L. L., Cheryl Wyn‐Evans, T. & Diener, E. Ontogeny of the murine immune system: development of antigen recognition and immune responsiveness. Eur. J. Immunol. 3, 224–228 (1973).
Pasquier, L. D. Ontogeny of the immune response in animals having less than one million lymphocytes: the larvae of the toad Alytes obstetricans. Immunology 19, 353 (1970).
Leighton, P. A., Morales, J., Harriman, W. D. & Ching, K. H. V(D)J rearrangement is dispensable for producing CDR-H3 sequence diversity in a gene converting species. Front. Immunol. 9, 362694 (2018).
Weber, J., Peng, H. & Rader, C. From rabbit antibody repertoires to rabbit monoclonal antibodies. Exp. Mol. Med. 49, e305–e305 (2017).
Hozumi, N. & Tonegawa, S. Evidence for somatic rearrangement of immunoglobulin genes coding for variable and constant regions. Proc. Natl Acad. Sci. USA 73, 3628–3632 (1976).
Rees, A. R. Understanding the human antibody repertoire. MAbs 12, 1729683 (2020).
Saada, R., Weinberger, M., Shahaf, G. & Mehr, R. Models for antigen receptor gene rearrangement: CDR3 length. Immunol. Cell Biol. 85, 323–332 (2007).
Collins, A. M. & Jackson, K. J. L. On being the right size: antibody repertoire formation in the mouse and human. Immunogenetics 70, 143–158 (2018).
Köhler, G. & Milstein, C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 256, 495–497 (1975).
Lieberman, R., Potter, M., Mushinski, E. B., Humphrey, W. & Rudikoff, S. Genetics of a new IgVH (T15 idiotype) marker in the mouse regulating natural antibody to phosphorylcholine. J. Exp. Med. 139, 983–1001 (1974).
Claflin, J. L. Uniformity in the clonal repertoire for the immune response to phosphorylcholine in mice. Eur. J. Immunol. 6, 669–674 (1976).
Winfield, J. B., Mage, R. G. & Alexander, C. B. Anti-p-azobenzenearsonate antibody of restricted heterogeneity. II. Idiotypes of antibodies produced during a 33-month period. J. Immunol. 110, 729–735 (1973).
Mage, R. G., Pincus, J. H., Alexander, C. & Freedman, M. H. Studies of hyperimmune restricted and partially restricted anti-pneumococcal polysaccharide antibodies from allotype-defined pedigreed rabbits. II. Allotypes and idiotypes in sera and electrofocused fractions. Eur. J. Immunol. 4, 560–564 (1974).
Briles, D. E. & Davie, J. M. Clonal dominance. I. Restricted nature of the IgM antibody response to group A streptococcal carbohydrate in mice. J. Exp. Med. 141, 1291–1307 (1975).
Jack, R. S., Imanishi‐Kari, T. & Rajewsky, K. Idiotypic analysis of the response of C57BL/6 mice to the (4-hydroxy-3-nitrophenyl)acetyl group. Eur. J. Immunol. 7, 559–565 (1977).
Barrett, D. J. & Ayoub, E. M. IgG2 subclass restriction of antibody to pneumococcal polysaccharides. Clin. Exp. Immunol. 63, 127 (1986).
Perlmutter, R. M., Hansburg, D., Briles, D. E., Nicolotti, R. A. & David, J. M. Subclass restriction of murine anti-carbohydrate antibodies. J. Immunol. 121, 566–572 (1978).
Briles, D. E., Forman, C., Hudak, S. & Claflin, J. L. The effects of subclass on the ability of anti-phosphocholine antibodies to protect mice from fatal infection with Streptococcus pneumoniae. J. Mol. Cell Immunol. 1, 305–309 (1984).
Yuan, R., Casadevall, A., Spira, G. & Scharff, M. D. Isotype switching from IgG3 to IgG1 converts a nonprotective murine antibody to Cryptococcus neoformans into a protective antibody. J. Immunol. 154, 1810–1816 (1995).
Abboud, N. et al. A requirement for FcγR in antibody-mediated bacterial toxin neutralization. J. Exp. Med. 207, 2395–2405 (2010).
Saggy, I. et al. Antibody isolation from immunized animals: comparison of phage display and antibody discovery via V gene repertoire mining. Protein Eng. Des. Sel. 25, 539–549 (2012).
Latham Claflin, J., Lieberman, R. & Davie, J. M. Clonal nature of the immune response to phosphorylcholine. I. Specificity, class, and idiotype of phosphorylcholine-binding receptors on lymphoid cells. J. Exp. Med. 139, 58–73 (1974).
Morahan, G., Berek, C. & Miller, J. F. A. P. An idiotypic determinant formed by both immunoglobulin constant and variable regions. Nature 301, 720–722 (1983).
Diamond, B. & Scharff, M. D. Somatic mutation of the T15 heavy chain gives rise to an antibody with autoantibody specificity. Proc. Natl Acad. Sci. USA 81, 5841–5844 (1984).
Kieber-Emmons, T. et al. The promise of the anti-idiotype concept. Front. Oncol. 2, 196 (2012).
De Bono, B., Madera, M. & Chothia, C. VH gene segments in the mouse and human genomes. J. Mol. Biol. 342, 131–143 (2004).
Chevillard, C., Ozaki, J., Herring, C. D. & Riblet, R. A three-megabase yeast artificial chromosome contig spanning the C57BL mouse Igh locus. J. Immunol. 168, 5659–5666 (2002).
Larijani, M. et al. The recombination difference between mouse κ and λ segments is mediated by a pair-wise regulation mechanism. Mol. Immunol. 43, 870–881 (2006).
Thiebe, R. et al. The variable genes and gene families of the mouse immunoglobulin κ locus. Eur. J. Immunol. 29, 2072–2081 (1999).
Mikocziova, I., Greiff, V. & Sollid, L. M. Immunoglobulin germline gene variation and its impact on human disease. Genes. Immun. 22, 205–217 (2021).
Li, H., Cui, X., Pramanik, S. & Chimge, N. O. Genetic diversity of the human immunoglobulin heavy chain VH region. Immunol. Rev. 190, 53–68 (2002).
Pallarès, N., Lefebvre, S., Contet, V., Matsuda, F. & Lefranc, M. P. The human immunoglobulin heavy variable genes. Exp. Clin. Immunogenet. 16, 36–60 (1999).
Glanville, J. et al. Naive antibody gene-segment frequencies are heritable and unaltered by chronic lymphocyte ablation. Proc. Natl Acad. Sci. USA 108, 20066–20071 (2011).
Boyd, S. D. et al. Individual variation in the germline Ig gene repertoire inferred from variable region gene rearrangements. J. Immunol. 184, 6986–6992 (2010).
Choi, N. M. et al. Deep sequencing of the murine Igh repertoire reveals complex regulation of nonrandom V gene rearrangement frequencies. J. Immunol. 191, 2393–2402 (2013).
DeKosky, B. J. et al. Large-scale sequence and structural comparisons of human naive and antigen-experienced antibody repertoires. Proc. Natl Acad. Sci. USA 113, E2636–E2645 (2016).
Janeway, C. A. The immune system evolved to discriminate infectious nonself from noninfectious self. Immunol. Today 13, 11–16 (1992).
Mukherjee, J., Casadevall, A. & Scharff, M. D. Molecular characterization of the humoral responses to Cryptococcus neoformans infection and glucuronoxylomannan-tetanus toxoid conjugate immunization. J. Exp. Med. 177, 1105–1116 (1993).
Casadevall, A. & Scharff, M. D. The mouse antibody response to infection with Cryptococcus neoformans: VH and VL usage in polysaccharide binding antibodies. J. Exp. Med. 174, 151–160 (1991).
Andersen, P. S. et al. Extensive restrictions in the VH sequence usage of the human antibody response against the Rhesus D antigen. Mol. Immunol. 44, 412–422 (2007).
Adderson, E. E., Shackelford, P. G., Quinn, A. & Carroll, W. L. Restricted Ig H chain V gene usage in the human antibody response to Haemophilus influenzae type b capsular polysaccharide. J. Immunol. 147, 1667–1674 (1991).
Solin, M. L., Kaartinen, M. & Mäkelä, O. The same few V genes account for a majority of oxazolone antibodies in most mouse strains. Mol. Immunol. 29, 1357–1362 (1992).
Kocher, H. P., Berek, C. & Jaton, J. C. The immune response of BALB/c mice to phosphorylcholine is restricted to a limited number of VH- and VL-isotypes. Mol. Immunol. 18, 1027–1033 (1981).
Wysocki, L. J., Gridley, T., Huang, S., Grandea, A. G. & Gefter, M. L. Single germline VH and V κ genes encode predominating antibody variable regions elicited in strain A mice by immunization with p-azophenylarsonate. J. Exp. Med. 166, 1–11 (1987).
Williams, J. V., Weitkamp, J. H., Blum, D. L., LaFleur, B. J. & Crowe, J. E. The human neonatal B cell response to respiratory syncytial virus uses a biased antibody variable gene repertoire that lacks somatic mutations. Mol. Immunol. 47, 407 (2009).
Gorny, M. K. et al. Preferential use of the VH5-51 gene segment by the human immune response to code for antibodies against the V3 domain of HIV-1. Mol. Immunol. 46, 917 (2009).
Robbiani, D. F. et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature 584, 437–442 (2020).
Wang, Y. et al. A large-scale systematic survey reveals recurring molecular features of public antibody responses to SARS-CoV-2. Immunity 55, 1105 (2022).
Barnes, C. O. et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature 588, 682–687 (2020).
Barnes, C. O. et al. Structures of human antibodies bound to SARS-CoV-2 spike reveal common epitopes and recurrent features of antibodies. Cell 182, 828 (2020).
Shrestha, L. B., Tedla, N. & Bull, R. A. Broadly-neutralizing antibodies against emerging SARS-CoV-2 variants. Front. Immunol. 12, 752003 (2021).
Dejnirattisai, W. et al. The antigenic anatomy of SARS-CoV-2 receptor binding domain. Cell 184, 2183 (2021).
Rapp, M. et al. Modular basis for potent SARS-CoV-2 neutralization by a prevalent VH1-2-derived antibody class. Cell Rep. 35, 108950 (2021).
Greenspan, N. S. & Di Cera, E. Defining epitopes: it’s not as easy as it seems. Nat. Biotechnol. 17, 936–937 (1999).
Hawa, M. I. et al. Antibodies to IA-2 and GAD65 in type 1 and type 2 diabetes: isotype restriction and polyclonality. Diabetes Care 23, 228–233 (2000).
Wang, N. et al. Conserved amino acid networks involved in antibody variable domain interactions. Proteins 76, 99–114 (2009).
Scott, M. G. & Fleischman, J. B. Preferential idiotype-isotype associations in antibodies to dinitrophenyl antigens. J. Immunol. 128, 2622–2628 (1982).
Abraham, K. M. & Teale, J. M. Isotype restriction during infection of mice with the cestode Mesocestoides corti: role of immune suppression. J. Immunol. 138, 1699–1704 (1987).
Khalife, J. et al. Isotypic restriction of the antibody response to human immunodeficiency virus. AIDS Res. Hum. Retroviruses 4, 3–9 (1988).
Pritsch, O. et al. Can isotype switch modulate antigen-binding affinity and influence clonal selection? Eur. J. Immunol. 30, 3387–3395 (2000).
Eryilmaz, E. et al. Global structures of IgG isotypes expressing identical variable regions. Mol. Immunol. 56, 588–598 (2013).
Casadevall, A. & Janda, A. Immunoglobulin isotype influences affinity and specificity. Proc. Natl Acad. Sci. USA 109, 12272–12273 (2012).
Morelock, M. M. et al. Isotype choice for chimeric antibodies affects binding properties. J. Biol. Chem. 269, 13048–13055 (1994).
Tudor, D. et al. Isotype modulates epitope specificity, affinity, and antiviral activities of anti-HIV-1 human broadly neutralizing 2F5 antibody. Proc. Natl Acad. Sci. USA 109, 12680–12685 (2012).
Porter, R. R. Separation and isolation of fractions of rabbit γ-globulin containing the antibody and antigenic combining sites. Nature 182, 670–671 (1958).
Wibmer, C. K., Moore, P. L. & Morris, L. HIV broadly neutralizing antibody targets. Curr. Opin. HIV AIDS 10, 135–143 (2015).
D’Angelo, S. et al. Many routes to an antibody heavy-chain CDR3: necessary, yet insufficient, for specific binding. Front. Immunol. 9, 395 (2018).
Vandyk, L. & Meek, K. Assembly of IgH CDR3: mechanism, regulation, and influence on antibody diversity. Int. Rev. Immunol. 8, 123–133 (1992).
Xu, J. L. & Davis, M. M. Diversity in the CDR3 region of VH is sufficient for most antibody specificities. Immunity 13, 37–45 (2000).
Davis, M. M. The evolutionary and structural ‘logic’ of antigen receptor diversity. Semin. Immunol. 16, 239–243 (2004).
Chothia, C. & Lesk, A. M. Canonical structures for the hypervariable regions of immunoglobulins. J. Mol. Biol. 196, 901–917 (1987).
Gorman, J. et al. Structure of super-potent antibody CAP256-VRC26.25 in complex with HIV-1 envelope reveals a combined mode of trimer-apex recognition. Cell Rep. 31, 107488 (2020).
Liu, L. et al. An antibody class with a common CDRH3 motif broadly neutralizes sarbecoviruses. Sci. Transl. Med. 14, eabn6859 (2022).
Liu, H. et al. Human antibodies to SARS-CoV-2 with a recurring YYDRxG motif retain binding and neutralization to variants of concern including Omicron. Commun. Biol. 5, 766 (2022).
Stanfield, R. L., Zemla, A., Wilson, I. A. & Rupp, B. Antibody elbow angles are influenced by their light chain class. J. Mol. Biol. 357, 1566–1574 (2006).
Lesk, A. M. & Chothia, C. Elbow motion in the immunoglobulins involves a molecular ball-and-socket joint. Nature 335, 188–190 (1988).
Chiu, M. L., Goulet, D. R., Teplyakov, A. & Gilliland, G. L. Antibody structure and function: the basis for engineering therapeutics. Antibodies 8, 55 (2019).
Toughiri, R. et al. Comparing domain interactions within antibody Fabs with κ and λ light chains. MAbs 8, 1276–1285 (2016).
Zhao, J., Zhang, B., Zhu, J., Nussinov, R. & Ma, B. Structure and energetic basis of overrepresented λ light chain in systemic light chain amyloidosis patients. Biochim. Biophys. Acta Mol. Basis Dis. 1864, 2294–2303 (2018).
Raybould, M. I. J., Turnbull, O. M., Suter, A., Guloglu, B. & Deane, C. M. Contextualising the developability risk of antibodies with λ light chains using enhanced therapeutic antibody profiling. Commun. Biol. 7, 62 (2024).
Fernández-Quintero, M. L. et al. Surprisingly fast interface and elbow angle dynamics of antigen-binding fragments. Front. Mol. Biosci. 7, 609088 (2020).
Knapp, B., Dunbar, J., Alcala, M. & Deane, C. M. Variable regions of antibodies and T-cell receptors may not be sufficient in molecular simulations investigating binding. J. Chem. Theory Comput. 13, 3097–3105 (2017).
McConnell, S. A. & Casadevall, A. Immunoglobulin constant regions provide stabilization to the paratope and enforce epitope specificity. J. Biol. Chem. 300, 107397 (2024).
Röthlisberger, D., Honegger, A. & Plückthun, A. Domain interactions in the Fab fragment: a comparative evaluation of the single-chain Fv and Fab format engineered with variable domains of different stability. J. Mol. Biol. 347, 773–789 (2005).
Vidarsson, G., Dekkers, G. & Rispens, T. IgG subclasses and allotypes: from structure to effector functions. Front. Immunol. 5, 117227 (2014).
Zhang, X. et al. 3D structural fluctuation of IgG1 antibody revealed by individual particle electron tomography. Sci. Rep. 5, 1–14 (2015).
Saphire, E. O., Parren, P. W. H. I., Barbas, C. F., Burton, D. R. & Wilson, I. A. Crystallization and preliminary structure determination of an intact human immunoglobulin, b12: an antibody that broadly neutralizes primary isolates of HIV-1. Acta Crystallogr. D Biol. Crystallogr. 57, 168–171 (2001).
Feige, M. J. et al. An unfolded CH1 domain controls the assembly and secretion of IgG antibodies. Mol. Cell 34, 569–579 (2009).
Jäger, M. & Plückthun, A. Folding and assembly of an antibody Fv fragment, a heterodimer stabilized by antigen. J. Mol. Biol. 285, 2005–2019 (1999).
Feige, M. J. et al. The structural analysis of shark IgNAR antibodies reveals evolutionary principles of immunoglobulins. Proc. Natl Acad. Sci. USA 111, 8155–8160 (2014).
Zhang, T. et al. Genetic removal of the CH1 exon enables the production of heavy chain-only IgG in mice. Front. Immunol. 9, 2202 (2018).
Al Qaraghuli, M. M., Kubiak-Ossowska, K., Ferro, V. A. & Mulheran, P. A. Antibody-protein binding and conformational changes: identifying allosteric signalling pathways to engineer a better effector response. Sci. Rep. 10, 13696 (2020).
Sela-Culang, I., Alon, S. & Ofran, Y. A systematic comparison of free and bound antibodies reveals binding-related conformational changes. J. Immunol. 189, 4890–4899 (2012).
Rowe, E. S. & Tanford, C. Equilibrium and kinetics of the denaturation of a homogeneous human immunoglobulin light chain. Biochemistry 12, 4822–4827 (1973).
Garber, E. & Demarest, S. J. A broad range of Fab stabilities within a host of therapeutic IgGs. Biochem. Biophys. Res. Commun. 355, 751–757 (2007).
Torres, M., Fernandez-Fuentes, N., Fiser, A. & Casadevall, A. Exchanging murine and human immunoglobulin constant chains affects the kinetics and thermodynamics of antigen binding and chimeric antibody autoreactivity. PLoS ONE 2, e1310 (2007).
Ma, B., Tsai, C. J., Haliloǧlu, T. & Nussinov, R. Dynamic allostery: linkers are not merely flexible. Structure 19, 907–917 (2011).
Yogo, R. et al. The Fab portion of immunoglobulin G contributes to its binding to Fcγ receptor III. Sci. Rep. 9, 11957 (2019).
Su, C. T. T., Lua, W. H., Ling, W. L. & Gan, S. K. E. Allosteric effects between the antibody constant and variable regions: a study of IgA Fc mutations on antigen binding. Antibodies 7, 20 (2018).
Lua, W. H. et al. The effects of antibody engineering CH and CL in trastuzumab and pertuzumab recombinant models: impact on antibody production and antigen-binding. Sci. Rep. 8, 718 (2018).
Zhao, J., Nussinov, R. & Ma, B. The allosteric effect in antibody-antigen recognition. Methods Mol. Biol. 2253, 175–183 (2021).
Janda, A., Bowen, A., Greenspan, N. S. & Casadevall, A. Ig constant region effects on variable region structure and function. Front. Microbiol. 7, 22 (2016).
Xia, Y., Janda, A., Eryilmaz, E., Casadevall, A. & Putterman, C. The constant region affects antigen binding of antibodies to DNA by altering secondary structure. Mol. Immunol. 56, 28–37 (2013).
Bowen, A. & Casadevall, A. Revisiting the immunoglobulin intramolecular signaling hypothesis. Trends Immunol. 37, 721–723 (2016).
Yang, D., Kroe-Barrett, R., Singh, S., Roberts, C. J. & Laue, T. M. IgG cooperativity — Is there allostery? Implications for antibody functions and therapeutic antibody development. MAbs 9, 1231 (2017).
Oda, M., Kozono, H., Morii, H. & Azuma, T. Evidence of allosteric conformational changes in the antibody constant region upon antigen binding. Int. Immunol. 15, 417–426 (2003).
Pellequer, J. L., Chen, S. W., Roberts, V. A., Tainer, J. A. & Getzoff, E. D. Unraveling the effect of changes in conformation and compactness at the antibody VL-VH interface upon antigen binding. J. Mol. Recognit. 12, 267–275 (1999).
Schroeder, H. W. & Cavacini, L. Structure and function of immunoglobulins. J. Allergy Clin. Immunol. 125, S41–S52 (2010).
Notkins, A. L. Polyreactivity of antibody molecules. Trends Immunol. 25, 174–179 (2004).
Kanyavuz, A., Marey-Jarossay, A., Lacroix-Desmazes, S. & Dimitrov, J. D. Breaking the law: unconventional strategies for antibody diversification. Nat. Rev. Immunol. 19, 355–368 (2019).
Prigent, J. et al. Scarcity of autoreactive human blood IgA+ memory B cells. Eur. J. Immunol. 46, 2340–2351 (2016).
Planchais, C. et al. Potent human broadly SARS-CoV-2-neutralizing IgA and IgG antibodies effective against Omicron BA.1 and BA.2. J. Exp. Med. 219, e20220638 (2022).
Torres, M., Fernández-Fuentes, N., Fiser, A. & Casadevall, A. The immunoglobulin heavy chain constant region affects kinetic and thermodynamic parameters of antibody variable region interactions with antigen. J. Biol. Chem. 282, 13917–13927 (2007).
Xia, Y. et al. The constant region contributes to the antigenic specificity and renal pathogenicity of murine anti-DNA antibodies. J. Autoimmun. 39, 398–411 (2012).
MacCallum, R. M., Martin, A. C. R. & Thornton, J. M. Antibody-antigen interactions: contact analysis and binding site topography. J. Mol. Biol. 262, 732–745 (1996).
Vajdos, F. F. et al. Comprehensive functional maps of the antigen-binding site of an anti-ErbB2 antibody obtained with shotgun scanning mutagenesis. J. Mol. Biol. 320, 415–428 (2002).
Queen, C. et al. A humanized antibody that binds to the interleukin 2 receptor. Proc. Natl Acad. Sci. USA 86, 10029–10033 (1989).
Lamminmäki, U. & Kankare, J. A. Crystal structure of a recombinant anti-estradiol Fab fragment in complex with 17β -estradiol. J. Biol. Chem. 276, 36687–36694 (2001).
Arnaout, R. et al. High-resolution description of antibody heavy-chain repertoires in humans. PLoS ONE 6, e22365 (2011).
Oostindie, S. C., Lazar, G. A., Schuurman, J. & Parren, P. W. H. I. Avidity in antibody effector functions and biotherapeutic drug design. Nat. Rev. Drug Discov. 21, 715–735 (2022).
Melo-Braga, M. N. et al. Unveiling the multifaceted landscape of N-glycosylation in antibody variable domains: insights and implications. Int. J. Biol. Macromol. 257, 128362 (2023).
van de Bovenkamp, F. S., Hafkenscheid, L., Rispens, T. & Rombouts, Y. The emerging importance of IgG Fab glycosylation in immunity. J. Immunol. 196, 1435–1441 (2016).
Gupta, S., Jiskoot, W., Schöneich, C. & Rathore, A. S. Oxidation and deamidation of monoclonal antibody products: potential impact on stability, biological activity, and efficacy. J. Pharm. Sci. 111, 903–918 (2022).
Zhong, X. & D’Antona, A. M. A potential antibody repertoire diversification mechanism through tyrosine sulfation for biotherapeutics engineering and production. Front. Immunol. 13, 1072702 (2022).
Wang, F. et al. Structural and functional characterization of glycosylation in an immunoglobulin G1 to Cryptococcus neoformans glucuronoxylomannan. Mol. Immunol. 43, 987–998 (2006).
Dimitrov, J. D. et al. A cryptic polyreactive antibody recognizes distinct clades of HIV-1 glycoprotein 120 by an identical binding mechanism. J. Biol. Chem. 289, 17767–17779 (2014).
Hadzhieva, M. et al. Mechanism and functional implications of the heme-induced binding promiscuity of IgE. Biochemistry 54, 2061–2072 (2015).
Zhou, T., Hamer, D. H., Hendrickson, W. A., Sattentau, Q. J. & Kwong, P. D. Interfacial metal and antibody recognition. Proc. Natl Acad. Sci. USA 102, 14575–14580 (2005).
Greenspan, N. S. & Cooper, L. J. Cooperative binding by mouse IgG3 antibodies: implications for functional affinity, effector function, and isotype restriction. Springer Semin. Immunopathol. 15, 271–291 (1993).
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Immunology thanks the anonymous reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
McConnell, S.A., Casadevall, A. New insights into antibody structure with implications for specificity, variable region restriction and isotype choice. Nat Rev Immunol 25, 621–632 (2025). https://doi.org/10.1038/s41577-025-01150-9
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41577-025-01150-9