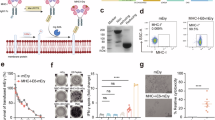
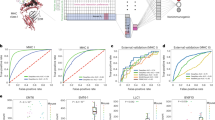
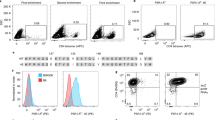
Abstract
MHC-E is a highly conserved, non-polymorphic MHC protein that engages inhibitory and activating receptors on natural killer (NK) cells and T cells and can also present antigens to T cell receptors. NK cell responses driven by activating receptor interactions with MHC-E are implicated in controlling chronic viral infections and cancer. Immunotherapeutic targeting of interactions between MHC-E and inhibitory receptors to increase the activation of NK cells and T cells shows promise in improving antitumour immune responses. Furthermore, MHC-E-restricted CD8+ T cells elicited by cytomegalovirus-based vaccines might, for certain infections and cancers, be more effective than CD8+ T cells restricted by classical MHC class I or class II molecules. The ability of MHC-E to regulate or mediate both innate and adaptive immune responses independently of the MHC haplotype of an individual raises the possibility of new, universally effective vaccines and immunotherapies for infectious disease and cancer. Although the therapeutic exploitation of MHC-E is still in its infancy, recent advances in the understanding of MHC-E biology show enormous potential, as described in this Review.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Trowsdale, J. & Knight, J. C. Major histocompatibility complex genomics and human disease. Annu. Rev. Genom. Hum. Genet. 14, 301–323 (2013).
D’Souza, M. P. et al. Casting a wider net: immunosurveillance by nonclassical MHC molecules. PLoS Pathog. 15, e1007567 (2019).
Braud, V. M. et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature 391, 795–799 (1998).
Borrego, F., Ulbrecht, M., Weiss, E. H., Coligan, J. E. & Brooks, A. G. Recognition of human histocompatibility leukocyte antigen (HLA)-E complexed with HLA class I signal sequence-derived peptides by CD94/NKG2 confers protection from natural killer cell-mediated lysis. J. Exp. Med. 187, 813–818 (1998).
Lee, N. et al. HLA-E is a major ligand for the natural killer inhibitory receptor CD94/NKG2A. Proc. Natl Acad. Sci. USA 95, 5199–5204 (1998). This seminal study, together with Braud et al. (1998) and Borrego et al. (1998), identifies the role of HLA-E binding to CD94–NKG2A and CD94–NKG2C in modulating NK cell activation according to levels of HLA-Ia signal sequence presentation on the target cell.
Aldrich, C. J. et al. Identification of a Tap-dependent leader peptide recognized by alloreactive T cells specific for a class Ib antigen. Cell 79, 649–658 (1994).
Lee, N., Goodlett, D. R., Ishitani, A., Marquardt, H. & Geraghty, D. E. HLA-E surface expression depends on binding of TAP-dependent peptides derived from certain HLA class I signal sequences. J. Immunol. 160, 4951–4960 (1998).
Braud, V., Jones, E. Y. & McMichael, A. The human major histocompatibility complex class Ib molecule HLA-E binds signal sequence-derived peptides with primary anchor residues at positions 2 and 9. Eur. J. Immunol. 27, 1164–1169 (1997).
Knapp, L. A., Cadavid, L. F. & Watkins, D. I. The MHC-E locus is the most well conserved of all known primate class I histocompatibility genes. J. Immunol. 160, 189–196 (1998).
Geraghty, D. E., Stockschleader, M., Ishitani, A. & Hansen, J. A. Polymorphism at the HLA-E locus predates most HLA-A and -B polymorphism. Hum. Immunol. 33, 174–184 (1992).
Strong, R. K. et al. HLA-E allelic variants. Correlating differential expression, peptide affinities, crystal structures, and thermal stabilities. J. Biol. Chem. 278, 5082–5090 (2003).
Gillespie, G. M., Quastel, M. N. & McMichael, A. J. HLA-E: immune receptor functional mechanisms revealed by structural studies. Immunol. Rev. 329, e13434 (2025). This work provides a comprehensive review of structural and biochemical studies of HLA-E.
van Hall, T., Oliveira, C. C., Joosten, S. A. & Ottenhoff, T. H. The other Janus face of Qa-1 and HLA-E: diverse peptide repertoires in times of stress. Microbes Infect. 12, 910–918 (2010).
Wei, X. H. & Orr, H. T. Differential expression of HLA-E, HLA-F, and HLA-G transcripts in human tissue. Hum. Immunol. 29, 131–142 (1990).
Middelburg, J. et al. The MHC-E peptide ligands for checkpoint CD94/NKG2A are governed by inflammatory signals, whereas LILRB1/2 receptors are peptide indifferent. Cell Rep. 42, 113516 (2023). This paper shows that MHC-E is a ligand for inhibitory receptors LIR1 and LIR2 irrespective of peptide ligand. Using CRISPR screening, the authors also identify gene products involved in processing of VL9 peptides.
Braud, V. M., Allan, D. S., Wilson, D. & McMichael, A. J. TAP- and tapasin-dependent HLA-E surface expression correlates with the binding of an MHC class I leader peptide. Curr. Biol. 8, 1–10 (1998).
He, W. et al. Intracellular trafficking of HLA-E and its regulation. J. Exp. Med. 220, e20221941 (2023). This study shows that the cytoplasmic tail of HLA-E contains internalization signals, which explains the short half-life of HLA-E on the cell surface.
Phillips, J. H. et al. CD94 and a novel associated protein (94AP) form a NK cell receptor involved in the recognition of HLA-A, HLA-B, and HLA-C allotypes. Immunity 5, 163–172 (1996).
Le Drean, E. et al. Inhibition of antigen-induced T cell response and antibody-induced NK cell cytotoxicity by NKG2A: association of NKG2A with SHP-1 and SHP-2 protein-tyrosine phosphatases. Eur. J. Immunol. 28, 264–276 (1998).
Carretero, M. et al. Specific engagement of the CD94/NKG2-A killer inhibitory receptor by the HLA-E class Ib molecule induces SHP-1 phosphatase recruitment to tyrosine-phosphorylated NKG2-A: evidence for receptor function in heterologous transfectants. Eur. J. Immunol. 28, 1280–1291 (1998).
Lanier, L. L., Corliss, B., Wu, J. & Phillips, J. H. Association of DAP12 with activating CD94/NKG2C NK cell receptors. Immunity 8, 693–701 (1998).
Horowitz, A. et al. Genetic and environmental determinants of human NK cell diversity revealed by mass cytometry. Sci. Transl. Med. 5, 208ra145 (2013).
Mingari, M. C. et al. HLA class I-specific inhibitory receptors in human T lymphocytes: interleukin 15-induced expression of CD94/NKG2A in superantigen- or alloantigen-activated CD8+ T cells. Proc. Natl Acad. Sci. USA 95, 1172–1177 (1998).
Andre, P. et al. Anti-NKG2A mAb is a checkpoint inhibitor that promotes anti-tumor immunity by unleashing both T and NK cells. Cell 175, 1731–1743.e13 (2018). This study shows that HLA-E–NKG2A is a new immune checkpoint for cancer therapy because blocking NKG2A increases the anti-tumour activity of NK cells and of tumour-specific T cells, including those induced by cancer vaccines.
Kaiser, B. K., Pizarro, J. C., Kerns, J. & Strong, R. K. Structural basis for NKG2A/CD94 recognition of HLA-E. Proc. Natl Acad. Sci. USA 105, 6696–6701 (2008).
Kaiser, B. K. et al. Interactions between NKG2x immunoreceptors and HLA-E ligands display overlapping affinities and thermodynamics. J. Immunol. 174, 2878–2884 (2005).
MacLachlan, B. J., Sullivan, L. C., Brooks, A. G., Rossjohn, J. & Vivian, J. P. Structure of the murine CD94-NKG2A receptor in complex with Qa-1b presenting an MHC-I leader peptide. FEBS J. 291, 1530–1544 (2024). This paper shows that the high-resolution structure of CD94–NKG2A with the murine MHC-E homologue Qa-1b shows similarities and differences compared with their human counterparts, thus paving the way for a humanized mouse.
Michaëlsson, J. et al. A signal peptide derived from hsp60 binds HLA-E and interferes with CD94/NKG2A recognition. J. Exp. Med. 196, 1403–1414 (2002).
Huisman, B. D. et al. High-throughput characterization of HLA-E-presented CD94/NKG2x ligands reveals peptides which modulate NK cell activation. Nat. Commun. 14, 4809 (2023). This work has developed a systematic screen of peptides binding to HLA-E and their effect on receptor binding that reveals differential peptide specificities of CD94–NKG2A and CD94–NKG2C.
Willcox, B. E., Thomas, L. M. & Bjorkman, P. J. Crystal structure of HLA-A2 bound to LIR-1, a host and viral major histocompatibility complex receptor. Nat. Immunol. 4, 913–919 (2003).
Shiroishi, M. et al. Human inhibitory receptors Ig-like transcript 2 (ILT2) and ILT4 compete with CD8 for MHC class I binding and bind preferentially to HLA-G. Proc. Natl Acad. Sci. USA 100, 8856–8861 (2003).
Horowitz, A. et al. Class I HLA haplotypes form two schools that educate NK cells in different ways. Sci. Immunol. 1, eaag1672 (2016).
Lin, Z. et al. HLA class I signal peptide polymorphism determines the level of CD94/NKG2-HLA-E-mediated regulation of effector cell responses. Nat. Immunol. 24, 1087–1097 (2023). This work shows that VL9 sequence polymorphism in HLA alleles is reflected in HCMV UL40 VL9 and impacts NK cell responses.
Cazzetta, V., Depierreux, D., Colucci, F., Mikulak, J. & Mavilio, D. NKG2A immune checkpoint in Vδ2 T cells: emerging application in cancer immunotherapy. Cancers 15, 1264 (2023).
Rapaport, A. S. et al. The inhibitory receptor NKG2A sustains virus-specific CD8+ T cells in response to a lethal poxvirus infection. Immunity 43, 1112–1124 (2015).
Zhang, X., Feng, J., Chen, S., Yang, H. & Dong, Z. Synergized regulation of NK cell education by NKG2A and specific Ly49 family members. Nat. Commun. 10, 5010 (2019).
Kaulfuss, M. et al. The NK cell checkpoint NKG2A maintains expansion capacity of human NK cells. Sci. Rep. 13, 10555 (2023).
Siemaszko, J., Marzec-Przyszlak, A. & Bogunia-Kubik, K. Activating NKG2C receptor: functional characteristics and current strategies in clinical applications. Arch. Immunol. Ther. Exp. 71, 9 (2023).
Lauterbach, N., Wieten, L., Popeijus, H. E., Voorter, C. E. & Tilanus, M. G. HLA-E regulates NKG2C+ natural killer cell function through presentation of a restricted peptide repertoire. Hum. Immunol. 76, 578–586 (2015).
Rolle, A., Meyer, M., Calderazzo, S., Jager, D. & Momburg, F. Distinct HLA-E peptide complexes modify antibody-driven effector functions of adaptive NK cells. Cell Rep. 24, 1967–1976.e4 (2018). This study shows that the HLA-E ligandome affects the activity and proliferation of NKG2C+ NK cells.
Lopez-Verges, S. et al. Expansion of a unique CD57+NKG2Chi natural killer cell subset during acute human cytomegalovirus infection. Proc. Natl Acad. Sci. USA 108, 14725–14732 (2011).
Foley, B. et al. Cytomegalovirus reactivation after allogeneic transplantation promotes a lasting increase in educated NKG2C+ natural killer cells with potent function. Blood 119, 2665–2674 (2012).
Lopez-Botet, M., De Maria, A., Muntasell, A., Della Chiesa, M. & Vilches, C. Adaptive NK cell response to human cytomegalovirus: facts and open issues. Semin. Immunol. 65, 101706 (2023). This paper provides a comprehensive review of HCMV-induced adaptive NK cells.
Della Chiesa, M. et al. Human cytomegalovirus infection promotes rapid maturation of NK cells expressing activating killer Ig-like receptor in patients transplanted with NKG2C−/− umbilical cord blood. J. Immunol. 192, 1471–1479 (2014).
Luetke-Eversloh, M. et al. Human cytomegalovirus drives epigenetic imprinting of the IFNG locus in NKG2Chi natural killer cells. PLoS Pathog. 10, e1004441 (2014).
Lee, J. et al. Epigenetic modification and antibody-dependent expansion of memory-like NK cells in human cytomegalovirus-infected individuals. Immunity 42, 431–442 (2015).
Schlums, H. et al. Cytomegalovirus infection drives adaptive epigenetic diversification of NK cells with altered signaling and effector function. Immunity 42, 443–456 (2015).
Gao, F. et al. Biology and clinical relevance of HCMV-associated adaptive NK cells. Front. Immunol. 13, 830396 (2022).
Costa-Garcia, M. et al. Antibody-mediated response of NKG2Cbright NK cells against human cytomegalovirus. J. Immunol. 194, 2715–2724 (2015).
Ulbrecht, M. et al. Cutting edge: the human cytomegalovirus UL40 gene product contains a ligand for HLA-E and prevents NK cell-mediated lysis. J. Immunol. 164, 5019–5022 (2000).
Tomasec, P. et al. Surface expression of HLA-E, an inhibitor of natural killer cells, enhanced by human cytomegalovirus gpUL40. Science 287, 1031 (2000).
Powers, C. J. & Früh, K. Signal peptide-dependent inhibition of MHC class I heavy chain translation by rhesus cytomegalovirus. PLoS Pathog. 4, e1000150 (2008).
Brackenridge, S. et al. Regulation of the cell surface expression of classical and non-classical MHC proteins by the human cytomegalovirus UL40 and rhesus cytomegalovirus Rh67 proteins. J. Virol. 98, e0120624 (2024). This study shows that UL40 and Rh67 of HCMV and RhCMV, respectively, not only promote MHC-E egress to the cell surface via their VL9 sequences but also retain classical MHC-I proteins in the ER.
Hammer, Q. et al. Peptide-specific recognition of human cytomegalovirus strains controls adaptive natural killer cells. Nat. Immunol. 19, 453–463 (2018). This paper shows that VL9 sequences determine NKG2C-dependent NK cell activation.
Koldehoff, M., Ross, S. R., Dührsen, U., Beelen, D. W. & Elmaagacli, A. H. Early CMV-replication after allogeneic stem cell transplantation is associated with a reduced relapse risk in lymphoma. Leuk. Lymphoma 58, 822–833 (2017).
Ram, D. R. et al. Tracking KLRC2 (NKG2C)+ memory-like NK cells in SIV+ and rhCMV+ rhesus macaques. PLoS Pathog. 14, e1007104 (2018).
Truitt, L. L. et al. Impact of CMV infection on natural killer cell clonal repertoire in CMV-naive rhesus macaques. Front. Immunol. 10, 2381 (2019).
Martin Almazan, N. et al. Non-classical HLA-E restricted CMV 15-mer peptides are recognized by adaptive NK cells and induce memory responses. Front. Immunol. 14, 1230718 (2023).
Wilkinson, G. W. et al. Modulation of natural killer cells by human cytomegalovirus. J. Clin. Virol. 41, 206–212 (2008).
Redondo-Pachon, D. et al. Adaptive NKG2C+ NK cell response and the risk of cytomegalovirus infection in kidney transplant recipients. J. Immunol. 198, 94–101 (2017).
Ataya, M. et al. Long-term evolution of the adaptive NKG2C+ NK cell response to cytomegalovirus infection in kidney transplantation: an insight on the diversity of host-pathogen interaction. J. Immunol. 207, 1882–1890 (2021).
Vietzen, H. et al. Extent of cytomegalovirus replication in the human host depends on variations of the HLA-E/UL40 axis. mBio 12, e02996–02920 (2021).
Vietzen, H., Pollak, K., Honsig, C., Jaksch, P. & Puchhammer-Stockl, E. NKG2C deletion is a risk factor for human cytomegalovirus viremia and disease after lung transplantation. J. Infect. Dis. 217, 802–806 (2018).
Mittelbronn, M. et al. Elevated HLA-E levels in human glioblastomas but not in grade I to III astrocytomas correlate with infiltrating CD8+ cells. J. Neuroimmunol. 189, 50–58 (2007).
Gooden, M. et al. HLA-E expression by gynecological cancers restrains tumor-infiltrating CD8+ T lymphocytes. Proc. Natl Acad. Sci. USA 108, 10656–10661 (2011).
Benevolo, M. et al. High expression of HLA-E in colorectal carcinoma is associated with a favorable prognosis. J. Transl. Med. 9, 184 (2011).
Seliger, B. et al. HLA-E expression and its clinical relevance in human renal cell carcinoma. Oncotarget 7, 67360–67372 (2016).
Kamiya, T., Seow, S. V., Wong, D., Robinson, M. & Campana, D. Blocking expression of inhibitory receptor NKG2A overcomes tumor resistance to NK cells. J. Clin. Invest. 129, 2094–2106 (2019). This paper implicates NKG2A as a checkpoint for NK cell-mediated killing of tumour cells; see also Andre et al. (2018) and van Montfoort et al. (2018).
Iyer, R. F. et al. CD8+ T cell targeting of tumor antigens presented by HLA-E. Sci. Adv. 10, eadm7515 (2024). This work shows that RhCMV vectors expressing tumour-associated antigens elicit strong MHC-E-restricted T cell responses in non-human primates that cross-react with HLA-E-expressing human cancer cells.
Wen, M. et al. Expression and clinical significance of NKG2A and HLA-E in advanced laryngeal carcinoma. Pathol. Res. Pract. 260, 155383 (2024).
Liu, X. et al. Immune checkpoints HLA-E:CD94-NKG2A and HLA-C:KIR2DL1 complementarily shield circulating tumor cells from NK-mediated immune surveillance. Cell Discov. 10, 16 (2024).
Liu, X. et al. Immune checkpoint HLA-E:CD94-NKG2A mediates evasion of circulating tumor cells from NK cell surveillance. Cancer Cell 41, 272–287.e9 (2023).
van Hall, T. et al. Monalizumab: inhibiting the novel immune checkpoint NKG2A. J. Immunother. Cancer 7, 263 (2019).
Lin, A. & Yan, W. H. Heterogeneity of HLA-G expression in cancers: facing the challenges. Front. Immunol. 9, 2164 (2018).
Liu, W. et al. FcRγ gene editing reprograms conventional NK cells to display key features of adaptive human NK cells. iScience 23, 101709 (2020).
Lo Monaco, E. et al. Human leukocyte antigen E contributes to protect tumor cells from lysis by natural killer cells. Neoplasia 13, 822–830 (2011).
Green, M. L. et al. CMV reactivation after allogeneic HCT and relapse risk: evidence for early protection in acute myeloid leukemia. Blood 122, 1316–1324 (2013).
Elmaagacli, A. H. et al. Early human cytomegalovirus replication after transplantation is associated with a decreased relapse risk: evidence for a putative virus-versus-leukemia effect in acute myeloid leukemia patients. Blood 118, 1402–1412 (2011).
Cichocki, F. et al. Adaptive NK cell reconstitution is associated with better clinical outcomes. JCI Insight 4, e125553 (2019).
Litjens, N. H. R., van der Wagen, L., Kuball, J. & Kwekkeboom, J. Potential beneficial effects of cytomegalovirus infection after transplantation. Front. Immunol. 9, 389 (2018).
Chiu, E. et al. Anti-NKG2C/IL-15/anti-CD33 killer engager directs primary and iPSC-derived NKG2C+ NK cells to target myeloid leukemia. Mol. Ther. 29, 3410–3421 (2021).
Cho, H. et al. Adaptive natural killer cells facilitate effector functions of daratumumab in multiple myeloma. Clin. Cancer Res. 27, 2947–2958 (2021).
Bigley, A. B. et al. Latent cytomegalovirus infection enhances anti-tumour cytotoxicity through accumulation of NKG2C+ NK cells in healthy humans. Clin. Exp. Immunol. 185, 239–251 (2016).
Murad, S. et al. NKG2C+ NK cells for immunotherapy of glioblastoma multiforme. Int. J. Mol. Sci. 23, 5857 (2022).
Haroun-Izquierdo, A. et al. Adaptive single-KIR+NKG2C+ NK cells expanded from select superdonors show potent missing-self reactivity and efficiently control HLA-mismatched acute myeloid leukemia. J. Immunother. Cancer 10, e005577 (2022).
van Montfoort, N. et al. NKG2A blockade potentiates CD8 T cell immunity induced by cancer vaccines. Cell 175, 1744–1755.e15 (2018). Using mouse models of cancer, this study shows that cancer vaccines elicit NKG2A-expressing T cells that are less effective at controlling cancers unless NKG2A is blocked.
Sheffer, M. et al. Genome-scale screens identify factors regulating tumor cell responses to natural killer cells. Nat. Genet. 53, 1196–1206 (2021).
Li, Y. et al. Unlocking the therapeutic potential of the NKG2A-HLA-E immune checkpoint pathway in T cells and NK cells for cancer immunotherapy. J. Immunother. Cancer 12, e009934 (2024). This work is a recent review of ongoing clinical studies of NKG2A-targeting and HLA-E-targeting immune checkpoint inhibitors.
Fisher, J. G., Graham, L. V. & Blunt, M. D. Strategies to disrupt NKG2A:HLA-E interactions for improved anti-cancer immunity. Oncotarget 15, 501–503 (2024).
Borst, L. et al. NKG2A is a late immune checkpoint on CD8 T cells and marks repeated stimulation and cell division. Int. J. Cancer 150, 688–704 (2022).
Abd Hamid, M. et al. Enriched HLA-E and CD94/NKG2A interaction limits antitumor CD8+ tumor-infiltrating T lymphocyte responses. Cancer Immunol. Res. 7, 1293–1306 (2019).
Chen, Y. et al. CD8+ T cells form the predominant subset of NKG2A+ cells in human lung cancer. Front. Immunol. 10, 3002 (2019).
Fesneau, O. et al. IL-12 drives the expression of the inhibitory receptor NKG2A on human tumor-reactive CD8 T cells. Nat. Commun. 15, 9988 (2024).
Speiser, D. E. et al. In vivo expression of natural killer cell inhibitory receptors by human melanoma-specific cytolytic T lymphocytes. J. Exp. Med. 190, 775–782 (1999).
Ducoin, K. et al. Targeting NKG2A to boost anti-tumor CD8 T-cell responses in human colorectal cancer. Oncoimmunology 11, 2046931 (2022).
Salomé, B. et al. NKG2A and HLA-E define an alternative immune checkpoint axis in bladder cancer. Cancer Cell 40, 1027–1043.e9 (2022).
Battin, C. et al. NKG2A-checkpoint inhibition and its blockade critically depends on peptides presented by its ligand HLA-E. Immunology 166, 507–521 (2022).
Eugene, J. et al. The inhibitory receptor CD94/NKG2A on CD8+ tumor-infiltrating lymphocytes in colorectal cancer: a promising new druggable immune checkpoint in the context of HLAE/β2m overexpression. Mod. Pathol. 33, 468–482 (2020).
Haanen, J. B. & Cerundolo, V. NKG2A, a new kid on the immune checkpoint block. Cell 175, 1720–1722 (2018).
Aggarwal, C. et al. Updated results from COAST, a phase 2 study of durvalumab (D) ± oleclumab (O) or monalizumab (M) in patients (pts) with stage III unresectable non-small cell lung cancer (uNSCLC). J. Clin. Oncol. 42, 8046 (2024).
Barlesi, F. et al. PACIFIC-9: phase III trial of durvalumab + oleclumab or monalizumab in unresectable stage III non-small-cell lung cancer. Future Oncol. 20, 2137–2147 (2024).
Spinosa, P. et al. Quantitative modeling predicts competitive advantages of a next generation anti-NKG2A monoclonal antibody over monalizumab for the treatment of cancer. CPT Pharmacomet. Syst. Pharmacol. 10, 220–229 (2021).
Zhou, Y. et al. Generation, characterization, and preclinical studies of a novel NKG2A-targeted antibody BRY805 for cancer immunotherapy. Antibodies 13, 93 (2024).
Ravindranath, M. H., Filippone, E. J., Devarajan, A. & Asgharzadeh, S. Enhancing natural killer and CD8+ T cell-mediated anticancer cytotoxicity and proliferation of CD8+ T cells with HLA-E monospecific monoclonal antibodies. Monoclon. Antib. Immunodiagn. Immunother. 38, 38–59 (2019).
Hwang, J. K. et al. HLA-E-VL9 antibodies enhance NK cell and CD8+ T cell cytotoxicity against HIV-infected CD4+ T cells. Preprint at bioRxiv https://doi.org/10.1101/2024.07.08.602401 (2024).
Carlsten, M. et al. Bortezomib sensitizes multiple myeloma to NK cells via ER-stress-induced suppression of HLA-E and upregulation of DR5. Oncoimmunology 8, e1534664 (2019).
Tsao, H.-W. et al. Targeting the aminopeptidase ERAP enhances antitumor immunity by disrupting the NKG2A-HLA-E inhibitory checkpoint. Immunity 57, 2863–2878 (2024). This study shows the utility of targeting ERAP (which is known to be required for VL9 processing for presentation by HLA-E) as a strategy for inhibiting HLA-E–NKG2A-mediated checkpoint control of NK cell and CD8+ T cell activity.
Fisher, J. G. et al. XPO1 inhibition sensitises CLL cells to NK cell mediated cytotoxicity and overcomes HLA-E expression. Leukemia 37, 2036–2049 (2023).
Westin, J. R. et al. Efficacy and safety of CD19-directed CAR-T cell therapies in patients with relapsed/refractory aggressive B-cell lymphomas: observations from the JULIET, ZUMA-1, and TRANSCEND trials. Am. J. Hematol. 96, 1295–1312 (2021).
Hong, M. & Chen, Y. Y. Killer fatigue: transition to NK-cell-like phenotype is a signature of CAR-T cell exhaustion. Cell 184, 6017–6019 (2021).
Good, C. R. et al. An NK-like CAR T cell transition in CAR T cell dysfunction. Cell 184, 6081–6100.e26 (2021).
Myers, J. A. & Miller, J. S. Exploring the NK cell platform for cancer immunotherapy. Nat. Rev. Clin. Oncol. 18, 85–100 (2021).
Bexte, T. et al. CRISPR/Cas9 editing of NKG2A improves the efficacy of primary CD33-directed chimeric antigen receptor natural killer cells. Nat. Commun. 15, 8439 (2024). This paper shows that CAR NK cell activity against CD33-expressing target cells can be improved by gene editing to remove NKG2A expression.
Saetersmoen, M. et al. Targeting HLA-E-overexpressing cancers with a NKG2A/C switch receptor. Med 6, 100521 (2024). This paper shows that a fusion protein design of the NKG2A ligand-binding domain with the NKG2C signalling domain combines the increased affinity of NKG2A for VL9–HLA-E with the activating signalling provided by NKG2C.
Voogd, L. et al. Mtb-specific HLA-E-restricted T cells are induced during Mtb infection but not after BCG administration in non-human primates and humans. Vaccines 12, 1129 (2024).
Mazzarino, P. et al. Identification of effector-memory CMV-specific T lymphocytes that kill CMV-infected target cells in an HLA-E-restricted fashion. Eur. J. Immunol. 35, 3240–3247 (2005).
Garcia, P. et al. Human T cell receptor-mediated recognition of HLA-E. Eur. J. Immunol. 32, 936–944 (2002).
Pietra, G. et al. HLA-E-restricted recognition of cytomegalovirus-derived peptides by human CD8+ cytolytic T lymphocytes. Proc. Natl Acad. Sci. USA 100, 10896–10901 (2003).
Romagnani, C. et al. HLA-E-restricted recognition of human cytomegalovirus by a subset of cytolytic T lymphocytes. Hum. Immunol. 65, 437–445 (2004).
Jouand, N. et al. HCMV triggers frequent and persistent UL40-specific unconventional HLA-E-restricted CD8 T-cell responses with potential autologous and allogeneic peptide recognition. PLoS Pathog. 14, e1007041 (2018). This study shows that UL40-encoded VL9 sequences can be targeted by HLA-E-restricted T cells in HCMV-infected humans.
Rousseliere, A., Delbos, L., Bressollette, C., Berthaume, M. & Charreau, B. Mapping and characterization of HCMV-specific unconventional HLA-E-restricted CD8 T cell populations and associated NK and T cell responses using HLA/peptide tetramers and spectral flow cytometry. Int. J. Mol. Sci. 23, 263 (2021).
Allard, M. et al. HLA-E-restricted cross-recognition of allogeneic endothelial cells by CMV-associated CD8 T cells: a potential risk factor following transplantation. PLoS ONE 7, e50951 (2012).
Sullivan, L. C. et al. The presence of HLA-E-restricted, CMV-specific CD8+ T cells in the blood of lung transplant recipients correlates with chronic allograft rejection. PLoS ONE 10, e0135972 (2015).
Waters, S. et al. Do variations in the HLA-E ligand encoded by UL40 distinguish individuals susceptible to HCMV disease? Hum. Immunol. 84, 75–79 (2022).
Tarrago, D., Gonzalez, I. & Gonzalez-Escribano, M. F. HLA-E restricted cytomegalovirus UL40 peptide polymorphism may represent a risk factor following congenital infection. BMC Genom. 23, 455 (2022).
Sullivan, L. C. et al. Natural killer cell receptors regulate responses of HLA-E-restricted T cells. Sci. Immunol. 6, eabe9057 (2021).
Sullivan, L. C. et al. A conserved energetic footprint underpins recognition of human leukocyte antigen-E by two distinct αβ T cell receptors. J. Biol. Chem. 292, 21149–21158 (2017).
Hoare, H. L. et al. Structural basis for a major histocompatibility complex class Ib-restricted T cell response. Nat. Immunol. 7, 256–264 (2006).
Lo Monaco, E. et al. HLA-E: strong association with β2-microglobulin and surface expression in the absence of HLA class I signal sequence-derived peptides. J. Immunol. 181, 5442–5450 (2008).
Brackenridge, S., John, N., Früh, K., Borrow, P. & McMichael, A. J. The antibodies 3D12 and 4D12 recognise distinct epitopes and conformations of HLA-E. Front. Immunol. 15, 1329032 (2024).
Walters, L. C. et al. Pathogen-derived HLA-E bound epitopes reveal broad primary anchor pocket tolerability and conformationally malleable peptide binding. Nat. Commun. 9, 3137 (2018). This paper shows that HLA-E can bind a wide variety of peptides, and it suggests that some of the peptides targeted by CD8+ T cells bind very poorly to HLA-E.
Grant, E. J. et al. The unconventional role of HLA-E: the road less traveled. Mol. Immunol. 120, 101–112 (2020).
Pietra, G., Romagnani, C., Manzini, C., Moretta, L. & Mingari, M. C. The emerging role of HLA-E-restricted CD8+ T lymphocytes in the adaptive immune response to pathogens and tumors. J. Biomed. Biotechnol. 2010, 907092 (2010).
Lampen, M. H. et al. Alternative peptide repertoire of HLA-E reveals a binding motif that is strikingly similar to HLA-A2. Mol. Immunol. 53, 126–131 (2013).
McMurtrey, C. et al. T cell recognition of Mycobacterium tuberculosis peptides presented by HLA-E derived from infected human cells. PLoS ONE 12, e0188288 (2017).
Ruibal, P. et al. Peptide binding to HLA-E molecules in humans, nonhuman primates, and mice reveals unique binding peptides but remarkably conserved anchor residues. J. Immunol. 205, 2861–2872 (2020).
Celik, A. A., Kraemer, T., Huyton, T., Blasczyk, R. & Bade-Doding, C. The diversity of the HLA-E-restricted peptide repertoire explains the immunological impact of the Arg107Gly mismatch. Immunogenetics 68, 29–41 (2016).
Kraemer, T. et al. HLA-E: presentation of a broader peptide repertoire impacts the cellular immune response-implications on HSCT outcome. Stem Cell Int. 2015, 346714 (2015).
Weitzen, M., Shahbazy, M., Kapoor, S. & Caron, E. Deciphering the HLA-E immunopeptidome with mass spectrometry: an opportunity for universal mRNA vaccines and T-cell-directed immunotherapies. Front. Immunol. 15, 1442783 (2024).
van Hall, T. et al. The varicellovirus-encoded TAP inhibitor UL49.5 regulates the presentation of CTL epitopes by Qa-1b1. J. Immunol. 178, 657–662 (2007).
Guan, J., Peske, J. D., Taylor, J. A. & Shastri, N. The nonclassical immune surveillance for ERAAP function. Curr. Opin. Immunol. 70, 105–111 (2021).
Nagarajan, N. A., Gonzalez, F. & Shastri, N. Nonclassical MHC class Ib-restricted cytotoxic T cells monitor antigen processing in the endoplasmic reticulum. Nat. Immunol. 13, 579–586 (2012).
Doorduijn, E. M. et al. T cells engaging the conserved MHC class Ib molecule Qa-1b with TAP-independent peptides are semi-invariant lymphocytes. Front. Immunol. 9, 60 (2018).
Manoharan Valerio, M. et al. The promiscuous development of an unconventional Qa1b-restricted T cell population. Front. Immunol. 14, 1250316 (2023).
Geiger, K. M. et al. Murine cytomegalovirus downregulates ERAAP and induces an unconventional T cell response to self. Cell Rep. 42, 112317 (2023). This paper shows that downregulation of ERAAP by murine CMV results in the induction of T cells specific for Qa-1b presenting a similar self-peptide. The same Qa-1b-specific T cell population was observed in ERAAP-knockout mice by Nagarajan et al. (2012).
Picker, L. J., Lifson, J. D., Gale, M. Jr., Hansen, S. G. & Früh, K. Programming cytomegalovirus as an HIV vaccine. Trends Immunol. 44, 287–304 (2023). This work provides a comprehensive review on the ability of CMV vectors to elicit MHC-E-restricted T cells.
Walker, B. D. CMV, MHC-E, and the quest for an unconventional AIDS vaccine. Sci. Immunol. 6, eabi5830 (2021).
Barry, P. A. et al. Cytomegalovirus-vectored vaccines for HIV and other pathogens. AIDS 34, 335–349 (2020).
Früh, K. & Picker, L. CD8+ T cell programming by cytomegalovirus vectors: applications in prophylactic and therapeutic vaccination. Curr. Opin. Immunol. 47, 52–56 (2017).
Sylwester, A. W. et al. Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. J. Exp. Med. 202, 673–685 (2005).
Kern, F. et al. Distribution of human CMV-specific memory T cells among the CD8pos. subsets defined by CD57, CD27, and CD45 isoforms. Eur. J. Immunol. 29, 2908–2915 (1999).
Masopust, D. & Picker, L. J. Hidden memories: frontline memory T cells and early pathogen interception. J. Immunol. 188, 5811–5817 (2012).
Hansen, S. G. et al. Effector memory T cell responses are associated with protection of rhesus monkeys from mucosal simian immunodeficiency virus challenge. Nat. Med. 15, 293–299 (2009).
Hansen, S. G. et al. Addendum: Effector memory T cell responses are associated with protection of rhesus monkeys from mucosal simian immunodeficiency virus challenge. Nat. Med. 17, 1692 (2011).
Hansen, S. G. et al. Profound early control of highly pathogenic SIV by an effector memory T-cell vaccine. Nature 473, 523–527 (2011).
Hansen, S. G. et al. Immune clearance of highly pathogenic SIV infection. Nature 502, 100–104 (2013).
Hansen, S. G. et al. A live-attenuated RhCMV/SIV vaccine shows long-term efficacy against heterologous SIV challenge. Sci. Transl. Med. 11, eaaw2607 (2019).
Hansen, S. G. et al. Broadly targeted CD8+ T cell responses restricted by major histocompatibility complex E. Science 351, 714–720 (2016).
Hansen, S. G. et al. Cytomegalovirus vectors violate CD8+ T cell epitope recognition paradigms. Science 340, 1237874 (2013).
Malouli, D. et al. Reevaluation of the coding potential and proteomic analysis of the BAC-derived rhesus cytomegalovirus strain 68-1. J. Virol. 86, 8959–8973 (2012).
Malouli, D. et al. Cytomegaloviral determinants of CD8+ T cell programming and RhCMV/SIV vaccine efficacy. Sci. Immunol. 6, eabg5413 (2021). This paper shows that multiple chemokine-like viral gene products independently prevent the induction of MHC-E-restricted T cells by CMV.
Malouli, D. et al. Cytomegalovirus-vaccine-induced unconventional T cell priming and control of SIV replication is conserved between primate species. Cell Host Microbe 30, 1207–1218.e7 (2022).
Malouli, D. et al. Human cytomegalovirus UL18 prevents priming of MHC-E- and MHC-II-restricted CD8+ T cells. Sci. Immunol. 9, eadp5216 (2024). This paper shows that HCMV UL18, a MHC-I-like LIR1 ligand, prevents the induction of MHC-E-restricted T cells when inserted into RhCMV.
Verweij, M. C. et al. Modulation of MHC-E transport by viral decoy ligands is required for RhCMV/SIV vaccine efficacy. Science 372, eabe9233 (2021). This study shows that viral VL9 peptides encoded in viral proteins are required for the induction of MHC-E-restricted T cells and that these T cells are required for protection against SIV.
Hansen, S. G. et al. Evasion of CD8+ T cells is critical for superinfection by cytomegalovirus. Science 328, 102–106 (2010).
Powers, C., DeFilippis, V., Malouli, D. & Früh, K. Cytomegalovirus immune evasion. Curr. Top. Microbiol. Immunol. 325, 333–359 (2008).
Hansen, S. G. et al. Myeloid cell tropism enables MHC-E-restricted CD8+ T cell priming and vaccine efficacy by the RhCMV/SIV vaccine. Sci. Immunol. 7, eabn9301 (2022).
Hansen, S. G. et al. Late gene expression-deficient cytomegalovirus vectors elicit conventional T cells that do not protect against SIV. JCI Insight 8, e164692 (2023).
Hansen, S. G. et al. Cytomegalovirus vectors expressing Plasmodium knowlesi antigens induce immune responses that delay parasitemia upon sporozoite challenge. PLoS ONE 14, e0210252 (2019).
Hansen, S. G. et al. Prevention of tuberculosis in rhesus macaques by a cytomegalovirus-based vaccine. Nat. Med. 24, 130–143 (2018).
Malouli, D. et al. Cytomegalovirus vaccine vector-induced effector memory CD4+ T cells protect cynomolgus macaques from lethal aerosolized heterologous avian influenza challenge. Nat. Commun. 15, 6007 (2024).
Burwitz, B. J. et al. MHC-E-restricted CD8+ T cells target hepatitis B virus-infected human hepatocytes. J. Immunol. 204, 2169–2176 (2020).
Wang, Y. et al. Ad5-nCoV vaccination could induce HLA-E restricted CD8+ T cell responses specific for epitopes on severe acute respiratory syndrome coronavirus 2 spike protein. Viruses 16, 5 (2023).
Hogan, M. J. et al. Cryptic MHC-E epitope from influenza elicits a potent cytolytic T cell response. Nat. Immunol. 24, 1933–1946 (2023).
Walters, L. C. et al. Primary and secondary functions of HLA-E are determined by stability and conformation of the peptide-bound complexes. Cell Rep. 39, 110959 (2022).
Walters, L. C., McMichael, A. J. & Gillespie, G. M. Detailed and atypical HLA-E peptide binding motifs revealed by a novel peptide exchange binding assay. Eur. J. Immunol. 50, 2075–2091 (2020).
Ruibal, P. et al. Discovery of HLA-E-presented epitopes: MHC-E/peptide binding and T-cell recognition. Methods Mol. Biol. 2574, 15–30 (2022).
Yang, H. et al. HLA-E-restricted SARS-CoV-2-specific T cells from convalescent COVID-19 patients suppress virus replication despite HLA class Ia down-regulation. Sci. Immunol. 8, eabl8881 (2023). This study shows that HLA-E-restricted, SARS-CoV-2-specific CD8+ T cells can be isolated and expanded from individuals with COVID-19.
Yang, H. et al. HLA-E-restricted, Gag-specific CD8+ T cells can suppress HIV-1 infection, offering vaccine opportunities. Sci. Immunol. 6, eabg1703 (2021). This paper shows that HLA-E-restricted HIV-specific T cells can be isolated and expanded from HIV-naive individuals.
Ruibal, P. et al. Identification of HLA-E binding Mycobacterium tuberculosis-derived epitopes through improved prediction models. J. Immunol. 209, 1555–1565 (2022).
Paterson, R. L. et al. An HLA-E-targeted TCR bispecific molecule redirects T cell immunity against Mycobacterium tuberculosis. Proc. Natl Acad. Sci. USA 121, e2318003121 (2024). This work determines that a bispecific antibody consisting of Mtb–HLA-E-specific TCR and anti-CD3 moieties can redirect T cells to M. tuberculosis-infected targeted cells.
Wallace, Z. et al. Instability of the HLA-E peptidome of HIV presents a major barrier to therapeutic targeting. Mol. Ther. 32, 678–688 (2024).
Lemberg, M. K., Bland, F. A., Weihofen, A., Braud, V. M. & Martoglio, B. Intramembrane proteolysis of signal peptides: an essential step in the generation of HLA-E epitopes. J. Immunol. 167, 6441–6446 (2001).
Weihofen, A., Lemberg, M. K., Ploegh, H. L., Bogyo, M. & Martoglio, B. Release of signal peptide fragments into the cytosol requires cleavage in the transmembrane region by a protease activity that is specifically blocked by a novel cysteine protease inhibitor. J. Biol. Chem. 275, 30951–30956 (2000).
Bland, F. A., Lemberg, M. K., McMichael, A. J., Martoglio, B. & Braud, V. M. Requirement of the proteasome for the trimming of signal peptide-derived epitopes presented by the nonclassical major histocompatibility complex class I molecule HLA-E. J. Biol. Chem. 278, 33747–33752 (2003).
Coupel, S. et al. Expression and release of soluble HLA-E is an immunoregulatory feature of endothelial cell activation. Blood 109, 2806–2814 (2007).
Hengel, H. et al. A viral ER-resident glycoprotein inactivates the MHC-encoded peptide transporter. Immunity 6, 623–632 (1997).
Ahn, K. et al. The ER-luminal domain of the HCMV glycoprotein US6 inhibits peptide translocation by TAP. Immunity 6, 613–621 (1997).
Park, B. et al. Human cytomegalovirus inhibits tapasin-dependent peptide loading and optimization of the MHC class I peptide cargo for immune evasion. Immunity 20, 71–85 (2004).
Prod’homme, V. et al. Human cytomegalovirus UL40 signal peptide regulates cell surface expression of the NK cell ligands HLA-E and gpUL18. J. Immunol. 188, 2794–2804 (2012).
Davis, Z. B. et al. A conserved HIV-1-derived peptide presented by HLA-E renders infected T-cells highly susceptible to attack by NKG2A/CD94-bearing natural killer cells. PLoS Pathog. 12, e1005421 (2016).
Hannoun, Z. et al. Identification of novel HIV-1-derived HLA-E-binding peptides. Immunol. Lett. 202, 65–72 (2018).
Romero-Martin, L. et al. Disruption of the HLA-E/NKG2X axis is associated with uncontrolled HIV infections. Front. Immunol. 13, 1027855 (2022).
Peppa, D. et al. Adaptive reconfiguration of natural killer cells in HIV-1 infection. Front. Immunol. 9, 474 (2018).
Guma, M. et al. Human cytomegalovirus infection is associated with increased proportions of NK cells that express the CD94/NKG2C receptor in aviremic HIV-1-positive patients. J. Infect. Dis. 194, 38–41 (2006).
Mela, C. M. et al. Switch from inhibitory to activating NKG2 receptor expression in HIV-1 infection: lack of reversion with highly active antiretroviral therapy. AIDS 19, 1761–1769 (2005).
Mela, C. M. & Goodier, M. R. The contribution of cytomegalovirus to changes in NK cell receptor expression in HIV-1-infected individuals. J. Infect. Dis. 195, 158–159 (2007).
Maidji, E., Somsouk, M., Rivera, J. M., Hunt, P. W. & Stoddart, C. A. Replication of CMV in the gut of HIV-infected individuals and epithelial barrier dysfunction. PLoS Pathog. 13, e1006202 (2017).
Jost, S. et al. Antigen-specific memory NK cell responses against HIV and influenza use the NKG2/HLA-E axis. Sci. Immunol. 8, eadi3974 (2023).
Hasan, M. Z. et al. SARS-CoV-2 infection induces adaptive NK cell responses by spike protein-mediated induction of HLA-E expression. Emerg. Microbes Infect. 13, 2361019 (2024).
Hammer, Q. et al. SARS-CoV-2 Nsp13 encodes for an HLA-E-stabilizing peptide that abrogates inhibition of NKG2A-expressing NK cells. Cell Rep. 38, 110503 (2022).
Alrubayyi, A. et al. Natural killer cell responses during SARS-CoV-2 infection and vaccination in people living with HIV-1. Sci. Rep. 13, 18994 (2023).
Huot, N. et al. SARS-CoV-2 viral persistence in lung alveolar macrophages is controlled by IFN-γ and NK cells. Nat. Immunol. 24, 2068–2079 (2023).
Vietzen, H. et al. Torque teno viruses exhaust and imprint the human immune system via the HLA-E/NKG2A axis. Front. Immunol. 15, 1447980 (2024).
Vietzen, H. et al. HLA-E-restricted immune responses are crucial for the control of EBV infections and the prevention of PTLD. Blood 141, 1560–1573 (2022).
Vietzen, H., Hartenberger, S., Aberle, S. W. & Puchhammer-Stöckl, E. Dissection of the NKG2C NK cell response against Puumala orthohantavirus. PLoS Negl. Trop. Dis. 15, e0010006 (2021).
Antrobus, R. D. et al. Virus-specific cytotoxic T lymphocytes differentially express cell-surface leukocyte immunoglobulin-like receptor-1, an inhibitory receptor for class I major histocompatibility complex molecules. J. Infect. Dis. 191, 1842–1853 (2005).
Wu, H. L. et al. The role of MHC-E in T cell immunity is conserved among humans, rhesus macaques, and cynomolgus macaques. J. Immunol. 200, 49–60 (2018).
Picker, L. J., Hansen, S. G. & Lifson, J. D. New paradigms for HIV/AIDS vaccine development. Annu. Rev. Med. 63, 95–111 (2012).
Pfeiffer, T. et al. Posoleucel, an allogeneic, off-the-shelf multivirus-specific T-cell therapy, for the treatment of refractory viral infections in the post-HCT setting. Clin. Cancer Res. 29, 324–330 (2023).
Acknowledgements
The authors thank A. Townsend for the help with figures and graphics. The authors acknowledge funding by the National Institutes of Health: R01 AI175459 to K.F., P.B., G.M.G., A.J.M. and L.J.P.; P01AI174856 to K.F. and L.J.P.; P51 OD011092 to K.F. and L.J.P.; UM1 AI164567 to P.B., G.M.G. and A.J.M.; and UM1 AI144371 to P.B. and A.J.M.; by the National Institute for Health and Care Research: NIHR 203339 to P.B.; by the Wellcome Trust: 227388/Z/23/Z to G.M.G. and A.J.M.; and by the Gates Foundation: INV-055780 to G.M.G. and A.J.M.
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding authors
Ethics declarations
Competing interests
Oregon Health & Science University (OHSU), L.J.P. and K.F. have a substantial financial interest in Vir Biotechnology, Inc., a company that may have a commercial interest in the results of this research and technology. L.J.P. and K.F. are inventors of technology licensed to Vir Biotechnology. These potential individual and institutional conflicts of interest have been reviewed and managed by OHSU. A.J.M. and G.M.G. are inventors on HLA-E-related patents owned by Oxford University Innovation. A.J.M. and G.M.G. are cofounders of E-Biotherapy Ltd, a dormant company with a potential commercial interest in this research and technology.
Peer review
Peer review information
Nature Reviews Immunology thanks Michael Hogan, Amir Horowitz and Thorbald Van Hall for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Früh, K., Borrow, P., Gillespie, G.M. et al. Targeting MHC-E as a new strategy for vaccines and immunotherapeutics. Nat Rev Immunol (2025). https://doi.org/10.1038/s41577-025-01218-6
Accepted:
Published:
DOI: https://doi.org/10.1038/s41577-025-01218-6