Abstract
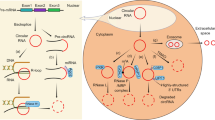
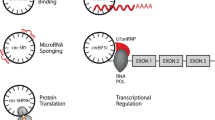
Circular RNA (circRNA) represents a type of RNA molecule characterized by a closed-loop structure that is distinct from linear RNA counterparts. Recent studies have revealed the emerging role of these circular transcripts in gene regulation and disease pathogenesis. However, their low expression levels and high sequence similarity to linear RNAs present substantial challenges for circRNA detection and characterization. Recent advances in long-read and single-cell RNA sequencing technologies, coupled with sophisticated deep learning-based algorithms, have revolutionized the investigation of circRNAs at unprecedented resolution and scale. This Review summarizes recent breakthroughs in circRNA discovery, characterization and functional analysis algorithms. We also discuss the challenges associated with integrating large-scale circRNA sequencing data and explore the potential future development of artificial intelligence (AI)-driven algorithms to unlock the full potential of circRNA research in biomedical applications.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Hansen, T. B. et al. Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–388 (2013).
Memczak, S. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495, 333–338 (2013).
Abdelmohsen, K. et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 14, 361–369 (2017).
Zhao, Q. et al. Targeting mitochondria-located circRNA SCAR alleviates NASH via reducing mROS output. Cell 183, 76–93 (2020).
Huang, D. et al. Tumour circular RNAs elicit anti-tumour immunity by encoding cryptic peptides. Nature 625, 593–602 (2024).
Liu, C. X. et al. Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell 177, 865–880 (2019).
Qu, L. et al. Circular RNA vaccines against SARS-CoV-2 and emerging variants. Cell 185, 1728–1744 (2022).
Liang, R. et al. Prime editing using CRISPR–Cas12a and circular RNAs in human cells. Nat. Biotechnol. 42, 1867–1875 (2024).
Yi, Z. et al. Engineered circular ADAR-recruiting RNAs increase the efficiency and fidelity of RNA editing in vitro and in vivo. Nat. Biotechnol. 40, 946–955 (2022).
Guo, S. K. et al. Therapeutic application of circular RNA aptamers in a mouse model of psoriasis. Nat. Biotechnol. 43, 236–246 (2025).
Feng, Z. et al. An in vitro-transcribed circular RNA targets the mitochondrial inner membrane cardiolipin to ablate EIF4G2+PTBP1+ pan-adenocarcinoma. Nat. Cancer 5, 30–46 (2024).
Xin, R. et al. isoCirc catalogs full-length circular RNA isoforms in human transcriptomes. Nat. Commun. 12, 266 (2021).
Liu, Z. et al. circFL-seq reveals full-length circular RNAs with rolling circular reverse transcription and nanopore sequencing. eLife 10, e69457 (2021).
Zhang, J. et al. Comprehensive profiling of circular RNAs with nanopore sequencing and CIRI-long. Nat. Biotechnol. 39, 836–845 (2021).
Wu, W., Zhang, J., Cao, X., Cai, Z. & Zhao, F. Exploring the cellular landscape of circular RNAs using full-length single-cell RNA sequencing. Nat. Commun. 13, 3242 (2022).
Zhou, Z. et al. CIRI-deep enables single-cell and spatial transcriptomic analysis of circular RNAs with deep learning. Adv. Sci. 11, e2308115 (2024).
Li, X. et al. A unified mechanism for intron and exon definition and back-splicing. Nature 573, 375–380 (2019).
Jeck, W. R. et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19, 141–157 (2013).
Conn, S. J. et al. The RNA binding protein Quaking regulates formation of circRNAs. Cell 160, 1125–1134 (2015).
You, X. & Conrad, T. O. Acfs: accurate circRNA identification and quantification from RNA-seq data. Sci. Rep. 6, 38820 (2016).
Gao, Y., Zhang, J. & Zhao, F. Circular RNA identification based on multiple seed matching. Brief. Bioinform. 19, 803–810 (2018).
Izuogu, O. G. et al. PTESFinder: a computational method to identify post-transcriptional exon shuffling (PTES) events. BMC Bioinformatics 17, 31 (2016).
Song, X. et al. Circular RNA profile in gliomas revealed by identification tool UROBORUS. Nucleic Acids Res. 44, e87 (2016).
Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at https://doi.org/10.48550/arXiv.1303.3997 (2013).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013).
Westholm, J. O. et al. Genome-wide analysis of Drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 9, 1966–1980 (2014).
Feng, J. et al. Genome-wide identification of cancer-specific alternative splicing in circRNA. Mol. Cancer 18, 35 (2019).
Cheng, J., Metge, F. & Dieterich, C. Specific identification and quantification of circular RNAs from sequencing data. Bioinformatics 32, 1094–1096 (2016).
Zhang, X. O. et al. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 26, 1277–1287 (2016).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Kim, D. & Salzberg, S. L. TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biol. 12, R72 (2011).
Hoffmann, S. et al. A multi-split mapping algorithm for circular RNA, splicing, trans-splicing and fusion detection. Genome Biol. 15, R34 (2014).
Wang, K. et al. MapSplice: accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res. 38, e178 (2010).
Kokot, M., Dehghannasiri, R., Baharav, T., Salzman, J. & Deorowicz, S. Scalable and unsupervised discovery from raw sequencing reads using SPLASH2. Nat. Biotechnol. https://doi.org/10.1038/s41587-024-02381-2 (2024).
Talhouarne, G. J. S. & Gall, J. G. Lariat intronic RNAs in the cytoplasm of vertebrate cells. Proc. Natl Acad. Sci. USA 115, E7970–E7977 (2018).
Lu, Z. et al. Metazoan tRNA introns generate stable circular RNAs in vivo. RNA 21, 1554–1565 (2015).
Ye, C. Y. et al. Full-length sequence assembly reveals circular RNAs with diverse non-GT/AG splicing signals in rice. RNA Biol. 14, 1055–1063 (2017).
Szabo, L. et al. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol. 16, 126 (2015).
Chuang, T. J. et al. NCLscan: accurate identification of non-co-linear transcripts (fusion, trans-splicing and circular RNA) with a good balance between sensitivity and precision. Nucleic Acids Res. 44, e29 (2016).
Zhang, J., Chen, S., Yang, J. & Zhao, F. Accurate quantification of circular RNAs identifies extensive circular isoform switching events. Nat. Commun. 11, 90 (2020).
Chen, C. Y. & Chuang, T. J. NCLcomparator: systematically post-screening non-co-linear transcripts (circular, trans-spliced, or fusion RNAs) identified from various detectors. BMC Bioinformatics 20, 3 (2019).
Gaffo, E., Buratin, A., Dal Molin, A. & Bortoluzzi, S. Sensitive, reliable and robust circRNA detection from RNA-seq with CirComPara2. Brief. Bioinform. 23, bbab418 (2022).
Vromman, M. et al. Large-scale benchmarking of circRNA detection tools reveals large differences in sensitivity but not in precision. Nat. Methods 20, 1159–1169 (2023).
Patro, R., Mount, S. M. & Kingsford, C. Sailfish enables alignment-free isoform quantification from RNA-seq reads using lightweight algorithms. Nat. Biotechnol. 32, 462–464 (2014).
Bray, N. L., Pimentel, H., Melsted, P. & Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527 (2016).
Li, M. et al. Quantifying circular RNA expression from RNA-seq data using model-based framework. Bioinformatics 33, 2131–2139 (2017).
Ashwal-Fluss, R. et al. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 56, 55–66 (2014).
Katz, Y., Wang, E. T., Airoldi, E. M. & Burge, C. B. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat. Methods 7, 1009–1015 (2010).
Ma, X. K. et al. CIRCexplorer3: a CLEAR pipeline for direct comparison of circular and linear RNA expression. Genomics Proteomics Bioinformatics 17, 511–521 (2019).
Morlion, A. et al. CiLiQuant: quantification of RNA junction reads based on their circular or linear transcript origin. Front. Bioinform. 2, 834034 (2022).
Enuka, Y. et al. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 44, 1370–1383 (2016).
Zhang, Y. et al. The biogenesis of nascent circular RNAs. Cell Rep. 15, 611–624 (2016).
Wagner, G. P., Kin, K. & Lynch, V. J. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 131, 281–285 (2012).
Salzman, J., Chen, R. E., Olsen, M. N., Wang, P. L. & Brown, P. O. Cell-type specific features of circular RNA expression. PLoS Genet. 9, e1003777 (2013).
Hulstaert, E. et al. Charting extracellular transcriptomes in the Human Biofluid RNA Atlas. Cell Rep. 33, 108552 (2020).
Liu, Z. et al. Detection of circular RNA expression and related quantitative trait loci in the human dorsolateral prefrontal cortex. Genome Biol. 20, 99 (2019).
Fay, M. P. Two-sided exact tests and matching confidence intervals for discrete data. R. J. 2, 53–58 (2010).
Feng, J. et al. GFOLD: a generalized fold change for ranking differentially expressed genes from RNA-seq data. Bioinformatics 28, 2782–2788 (2012).
Pamudurti, N. R. et al. circMbl functions in cis and in trans to regulate gene expression and physiology in a tissue-specific fashion. Cell Rep. 39, 110740 (2022).
Gao, Y. et al. Comprehensive identification of internal structure and alternative splicing events in circular RNAs. Nat. Commun. 7, 12060 (2016).
Hossain, M. T., Peng, Y., Feng, S. & Wei, Y. FcircSEC: an R package for full length circRNA sequence extraction and classification. Int. J. Genomics 2020, 9084901 (2020).
Abramson, J. et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630, 493–500 (2024).
Xiao, M. S. & Wilusz, J. E. An improved method for circular RNA purification using RNase R that efficiently removes linear RNAs containing G-quadruplexes or structured 3′ ends. Nucleic Acids Res. 47, 8755–8769 (2019).
Wu, W., Zhao, F. & Zhang, J. circAtlas 3.0: a gateway to 3 million curated vertebrate circular RNAs based on a standardized nomenclature scheme. Nucleic Acids Res. 52, D52–D60 (2024).
Zheng, Y., Ji, P., Chen, S., Hou, L. & Zhao, F. Reconstruction of full-length circular RNAs enables isoform-level quantification. Genome Med. 11, 2 (2019).
Huang, X. & Madan, A. CAP3: a DNA sequence assembly program. Genome Res. 9, 868–877 (1999).
Qin, Y. et al. Reference-free and de novo identification of circular RNAs. Preprint at bioRxiv https://doi.org/10.1101/2020.04.21.050617 (2020).
Peng, Y. et al. IDBA-tran: a more robust de novo de Bruijn graph assembler for transcriptomes with uneven expression levels. Bioinformatics 29, i326–i334 (2013).
Wu, J. et al. CircAST: full-length assembly and quantification of alternatively spliced isoforms in circular RNAs. Genomics Proteomics Bioinformatics 17, 522–534 (2019).
Wang, Y., Zhao, Y., Bollas, A., Wang, Y. & Au, K. F. Nanopore sequencing technology, bioinformatics and applications. Nat. Biotechnol. 39, 1348–1365 (2021).
Kelleher, C. D. & Champoux, J. J. Characterization of RNA strand displacement synthesis by Moloney murine leukemia virus reverse transcriptase. J. Biol. Chem. 273, 9976–9986 (1998).
Benson, G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27, 573–580 (1999).
Vaser, R., Sovic, I., Nagarajan, N. & Sikic, M. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 27, 737–746 (2017).
Gao, Y., Liu, B., Wang, Y. & Xing, Y. TideHunter: efficient and sensitive tandem repeat detection from noisy long-reads using seed-and-chain. Bioinformatics 35, i200–i207 (2019).
Rahimi, K., Veno, M. T., Dupont, D. M. & Kjems, J. Nanopore sequencing of brain-derived full-length circRNAs reveals circRNA-specific exon usage, intron retention and microexons. Nat. Commun. 12, 4825 (2021).
Unlu, I., Maguire, S., Guan, S. & Sun, Z. Induro-RT mediated circRNA-sequencing (IMCR-seq) enables comprehensive profiling of full-length and long circular RNAs from low input total RNA. Nucleic Acids Res. 52, e55 (2024).
Vincent, H. A. & Deutscher, M. P. Insights into how RNase R degrades structured RNA: analysis of the nuclease domain. J. Mol. Biol. 387, 570–583 (2009).
Szabo, L. & Salzman, J. Detecting circular RNAs: bioinformatic and experimental challenges. Nat. Rev. Genet. 17, 679–692 (2016).
Fuchs, S. et al. Generation of full-length circular RNA libraries for Oxford Nanopore long-read sequencing. PLoS ONE 17, e0273253 (2022).
Zhang, J. et al. Real-time and programmable transcriptome sequencing with PROFIT-seq. Nat. Cell Biol. 26, 2183–2194 (2024).
Ji, P. et al. Expanded expression landscape and prioritization of circular RNAs in mammals. Cell Rep. 26, 3444–3460 (2019).
Xia, S. et al. Comprehensive characterization of tissue-specific circular RNAs in the human and mouse genomes. Brief. Bioinform. 18, 984–992 (2017).
Nicolet, B. P. et al. Circular RNA expression in human hematopoietic cells is widespread and cell-type specific. Nucleic Acids Res. 46, 8168–8180 (2018).
Corces, M. R. et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat. Genet. 48, 1193–1203 (2016).
Garcia-Rodriguez, J. L. et al. Spatial profiling of circular RNAs in cancer reveals high expression in muscle and stromal cells. Cancer Res. 83, 3340–3353 (2023).
Kristensen, L. S. et al. Spatial expression analyses of the putative oncogene ciRS-7 in cancer reshape the microRNA sponge theory. Nat. Commun. 11, 4551 (2020).
Weng, W. et al. Circular RNA ciRS-7—a promising prognostic biomarker and a potential therapeutic target in colorectal cancer. Clin. Cancer Res. 23, 3918–3928 (2017).
He, R., Zhu, J., Ji, P. & Zhao, F. SEVtras delineates small extracellular vesicles at droplet resolution from single-cell transcriptomes. Nat. Methods 21, 259–266 (2024).
Dong, X. et al. Circular RNAs in the human brain are tailored to neuron identity and neuropsychiatric disease. Nat. Commun. 14, 5327 (2023).
Gaffo, E. et al. Circular RNA differential expression in blood cell populations and exploration of circRNA deregulation in pediatric acute lymphoblastic leukemia. Sci. Rep. 9, 14670 (2019).
Fan, X. et al. Single-cell RNA-seq transcriptome analysis of linear and circular RNAs in mouse preimplantation embryos. Genome Biol. 16, 148 (2015).
Dang, Y. et al. Tracing the expression of circular RNAs in human pre-implantation embryos. Genome Biol. 17, 130 (2016).
Verboom, K. et al. SMARTer single cell total RNA sequencing. Nucleic Acids Res. 47, e93 (2019).
Sheng, K., Cao, W., Niu, Y., Deng, Q. & Zong, C. Effective detection of variation in single-cell transcriptomes using MATQ-seq. Nat. Methods 14, 267–270 (2017).
Salmen, F. et al. High-throughput total RNA sequencing in single cells using VASA-seq. Nat. Biotechnol. 40, 1780–1793 (2022).
Xu, Z. et al. High-throughput single nucleus total RNA sequencing of formalin-fixed paraffin-embedded tissues by snRandom-seq. Nat. Commun. 14, 2734 (2023).
Isakova, A., Neff, N. & Quake, S. R. Single-cell quantification of a broad RNA spectrum reveals unique noncoding patterns associated with cell types and states. Proc. Natl Acad. Sci. USA 118, e2113568118 (2021).
Ruan, H. et al. Comprehensive characterization of circular RNAs in ~ 1000 human cancer cell lines. Genome Med. 11, 55 (2019).
Zhang, X. O. et al. Complementary sequence-mediated exon circularization. Cell 159, 134–147 (2014).
Liang, D. & Wilusz, J. E. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 28, 2233–2247 (2014).
Rogalska, M. E. et al. Transcriptome-wide splicing network reveals specialized regulatory functions of the core spliceosome. Science 386, 551–560 (2024).
Van Nostrand, E. L. et al. A large-scale binding and functional map of human RNA-binding proteins. Nature 583, 711–719 (2020).
Gonatopoulos-Pournatzis, T. et al. Genome-wide CRISPR–Cas9 interrogation of splicing networks reveals a mechanism for recognition of autism-misregulated neuronal microexons. Mol. Cell 72, 510–524 (2018).
Zhu, Y. et al. POSTAR2: deciphering the post-transcriptional regulatory logics. Nucleic Acids Res. 47, D203–D211 (2019).
Chen, Y. et al. CircNet 2.0: an updated database for exploring circular RNA regulatory networks in cancers. Nucleic Acids Res. 50, D93–D101 (2022).
Paz, I., Argoetti, A., Cohen, N., Even, N. & Mandel-Gutfreund, Y. RBPmap: a tool for mapping and predicting the binding sites of RNA-binding proteins considering the motif environment. Methods Mol. Biol. 2404, 53–65 (2022).
Riffo-Campos, A. L., Riquelme, I. & Brebi-Mieville, P. Tools for sequence-based miRNA target prediction: what to choose? Int. J. Mol. Sci. 17, 1987 (2016).
Chen, Y., Wang, J., Wang, C., Liu, M. & Zou, Q. Deep learning models for disease-associated circRNA prediction: a review. Brief. Bioinform. 23, bbac364 (2022).
Vromman, M., Vandesompele, J. & Volders, P. J. Closing the circle: current state and perspectives of circular RNA databases. Brief. Bioinform. 22, 288–297 (2021).
Wang, J. Z., Du, Z., Payattakool, R., Yu, P. S. & Chen, C. F. A new method to measure the semantic similarity of GO terms. Bioinformatics 23, 1274–1281 (2007).
van Laarhoven, T., Nabuurs, S. B. & Marchiori, E. Gaussian interaction profile kernels for predicting drug–target interaction. Bioinformatics 27, 3036–3043 (2011).
Fan, C., Lei, X. & Pan, Y. Prioritizing circRNA–disease associations with convolutional neural network based on multiple similarity feature fusion. Front. Genet. 11, 540751 (2020).
Deepthi, K. & Jereesh, A. S. An ensemble approach for circRNA–disease association prediction based on autoencoder and deep neural network. Gene 762, 145040 (2020).
Niu, M., Wang, C., Zhang, Z. & Zou, Q. A computational model of circRNA-associated diseases based on a graph neural network: prediction and case studies for follow-up experimental validation. BMC Biol. 22, 24 (2024).
Niu, M., Zou, Q. & Wang, C. GMNN2CD: identification of circRNA–disease associations based on variational inference and graph Markov neural networks. Bioinformatics 38, 2246–2253 (2022).
Wang, Y. et al. Collaborative deep learning improves disease-related circRNA prediction based on multi-source functional information. Brief. Bioinform. 24, bbad069 (2023).
Xu, C. & Zhang, J. Mammalian circular RNAs result largely from splicing errors. Cell Rep. 36, 109439 (2021).
Liu, C. X. & Chen, L. L. Circular RNAs: characterization, cellular roles, and applications. Cell 185, 2016–2034 (2022).
Jarlstad Olesen, M. T. & L, S. K. Circular RNAs as microRNA sponges: evidence and controversies. Essays Biochem. 65, 685–696 (2021).
Panda, A. C. et al. High-purity circular RNA isolation method (RPAD) reveals vast collection of intronic circRNAs. Nucleic Acids Res. 45, e116 (2017).
Abe, B. T., Wesselhoeft, R. A., Chen, R., Anderson, D. G. & Chang, H. Y. Circular RNA migration in agarose gel electrophoresis. Mol. Cell 82, 1768–1777 (2022).
Vromman, M. et al. Validation of circular RNAs using RT–qPCR after effective removal of linear RNAs by ribonuclease R. Curr. Protoc. 1, e181 (2021).
Vo, J. N. et al. The landscape of circular RNA in cancer. Cell 176, 869–881 (2019).
Hou, L., Zhang, J. & Zhao, F. Full-length circular RNA profiling by nanopore sequencing with CIRI-long. Nat. Protoc. 18, 1795–1813 (2023).
Chiang, T. W. et al. FL-circAS: an integrative resource and analysis for full-length sequences and alternative splicing of circular RNAs with nanopore sequencing. Nucleic Acids Res. 52, D115–D123 (2024).
Fuchs, S. et al. Defining the landscape of circular RNAs in neuroblastoma unveils a global suppressive function of MYCN. Nat. Commun. 14, 3936 (2023).
Lai, H. et al. exoRBase 2.0: an atlas of mRNA, lncRNA and circRNA in extracellular vesicles from human biofluids. Nucleic Acids Res. 50, D118–D128 (2022).
Zhao, J. et al. Circular RNA landscape in extracellular vesicles from human biofluids. Genome Med. 16, 126 (2024).
Rybak-Wolf, A. et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol. Cell 58, 870–885 (2015).
Jakobi, T., Uvarovskii, A. & Dieterich, C. circtools—a one-stop software solution for circular RNA research. Bioinformatics 35, 2326–2328 (2019).
Liu, X. et al. Identification of mecciRNAs and their roles in the mitochondrial entry of proteins. Sci. China Life Sci. 63, 1429–1449 (2020).
Li, J. H., Liu, S., Zhou, H., Qu, L. H. & Yang, J. H. starBase v2.0: decoding miRNA–ceRNA, miRNA–ncRNA and protein–RNA interaction networks from large-scale CLIP-seq data. Nucleic Acids Res. 42, D92–D97 (2014).
Kim, M. S. et al. A draft map of the human proteome. Nature 509, 575–581 (2014).
Yang, Q. et al. dbDEPC 3.0: the database of differentially expressed proteins in human cancer with multi-level annotation and drug indication. Database 2018, bay015 (2018).
Perez-Riverol, Y. et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 50, D543–D552 (2022).
Yang, Y. et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 27, 626–641 (2017).
Zhang, M. et al. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat. Commun. 9, 4475 (2018).
Ragan, C., Goodall, G. J., Shirokikh, N. E. & Preiss, T. Insights into the biogenesis and potential functions of exonic circular RNA. Sci. Rep. 9, 2048 (2019).
Floor, S. N. & Doudna, J. A. Tunable protein synthesis by transcript isoforms in human cells. eLife 5, e10921 (2016).
Liu, S., Zhu, A., He, C. & Chen, M. REPIC: a database for exploring the N6-methyladenosine methylome. Genome Biol. 21, 100 (2020).
Hansen, T. B. Signal and noise in circRNA translation. Methods 196, 68–73 (2021).
Huang, W. et al. TransCirc: an interactive database for translatable circular RNAs based on multi-omics evidence. Nucleic Acids Res. 49, D236–D242 (2021).
Zhu, J. et al. Custom microfluidic chip design enables cost-effective three-dimensional spatiotemporal transcriptomics with a wide field of view. Nat. Genet. 56, 2259–2270 (2024).
Hu, B. et al. High-resolution spatially resolved proteomics of complex tissues based on microfluidics and transfer learning. Cell 188, 734–748 (2025).
Long, Y. et al. Deciphering spatial domains from spatial multi-omics with SpatialGlue. Nat. Methods 21, 1658–1667 (2024).
Cao, Z. J. & Gao, G. Multi-omics single-cell data integration and regulatory inference with graph-linked embedding. Nat. Biotechnol. 40, 1458–1466 (2022).
Chu, Y. et al. A 5′ UTR language model for decoding untranslated regions of mRNA and function predictions. Nat. Mach. Intell. 6, 449–460 (2024).
Watson, J. L. et al. De novo design of protein structure and function with RFdiffusion. Nature 620, 1089–1100 (2023).
Cao, X., Cai, Z., Zhang, J. & Zhao, F. Engineering circular RNA medicines. Nat. Rev. Bioeng. https://doi.org/10.1038/s44222-024-00259-1 (2024).
Rettie, S. A. et al. Cyclic peptide structure prediction and design using AlphaFold. Preprint at bioRxiv https://doi.org/10.1101/2023.02.25.529956 (2023).
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (32130020 and 32025009 to F.Z. and 32200530 and 32422020 to J.Z.) and the National Key R&D Project (2021YFA1300500 and 2021YFA1302000 to J.Z.).
Author information
Authors and Affiliations
Contributions
F.Z. conceived the project. J.Z. conducted the initial literature research. All authors contributed to writing and edited the manuscript before submission.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Genetics thanks Jo Vandesompele and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Table 1
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, J., Zhao, F. Circular RNA discovery with emerging sequencing and deep learning technologies. Nat Genet 57, 1089–1102 (2025). https://doi.org/10.1038/s41588-025-02157-7
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41588-025-02157-7
This article is cited by
-
Circ_0008219 regulates proliferation and apoptosis in goat granulosa cells via a feedback loop with the transcription factor YY1
BMC Genomics (2026)
-
Comprehensive mapping of RNA modification dynamics and crosstalk via deep learning and nanopore direct RNA-sequencing
Nature Communications (2026)
-
Decoupling transcriptome layers: the distinct and variable nature of circular RNAs
BMC Biology (2025)
-
Detecting and quantifying circular RNAs in terabyte-scale RNA-seq datasets with CIRI3
Nature Biotechnology (2025)
-
Circular RNA profiling identifies circ_0001522, circ_0001278, and circ_0001801 as predictors of unfavorable prognosis and drivers of triple-negative breast cancer hallmarks
Cell Death Discovery (2025)