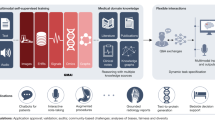
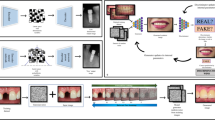
Abstract
Generative artificial intelligence (GAI) can automate a growing number of biomedical tasks, ranging from clinical decision support to design and analysis of research studies. GAI uses machine learning and transformer model architectures to generate useful text, images and sound data in response to user queries. While previous biomedical deep-learning applications have used general-purpose datasets and enormous volumes of labeled data for training, evidence now suggests that GAI models may perform better while requiring less training data—for example, using smaller, domain-specific datasets. Moreover, AI techniques have progressed from fully supervised training to less label-intensive approaches, such as weakly supervised or unsupervised fine-tuning and reinforcement learning. Recent iterations of GAI, such as agents, mixture-of-expert models and reasoning models, have further extended their capabilities to assist with complex and multistage tasks. Here, we provide an overview of recent technical advancements in GAI. We explore the potential of the latest generation of models to improve healthcare for clinicians and patients, and discuss validation approaches using specific examples to illustrate challenges and opportunities for further work.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Bengesi, S. et al. Advancements in generative AI: a comprehensive review of GANs, GPT, autoencoders, diffusion model, and transformers. IEEE Access 12, 69812–69837 (2024).
Sumner, J., Wang, Y., Tan, S. Y., Chew, E. H. H. & Wenjun Yip, A. Perspectives and experiences with large language models in health care: survey study. J. Med. Internet Res. 27, e67383 (2025).
Thirunavukarasu, A. J. et al. Large language models in medicine. Nat. Med. 29, 1930–1940 (2023).
Singhal, K. et al. Toward expert-level medical question answering with large language models. Nat. Med. 31, 943–950 (2025).
Amatriain, X. Transformer models: an introduction and catalog. Preprint at https://doi.org/10.48550/arXiv.2302.07730 (2023).
Tu, T. et al. Towards generalist biomedical AI. NEJM AI 1, AIoa2300138 (2024).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Abramson, J. et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630, 493–500 (2024).
Ren, F. et al. AlphaFold accelerates artificial intelligence powered drug discovery: efficient discovery of a novel CDK20 small molecule inhibitor. Chem. Sci. 14, 1443–1452 (2023).
DeepSeek-AI et al. DeepSeek-R1: incentivizing reasoning capability in LLMs via reinforcement learning. Preprint at https://doi.org/10.48550/arXiv.2501.12948 (2025).
Zou, J. & Topol, E. J. The rise of agentic AI teammates in medicine. Lancet 405, 457 (2025).
Thirunavukarasu, A. J. Large language models will not replace healthcare professionals: curbing popular fears and hype. J. R. Soc. Med. 116, 181–182 (2023).
Lee, P. The AI Revolution in Medicine: GPT-4 and Beyond (Pearson, 2023).
Lu, C. et al. The AI scientist: towards fully automated open-ended scientific discovery. Preprint at https://doi.org/10.48550/arXiv.2408.06292 (2024).
Esteva, A. et al. A guide to deep learning in healthcare. Nat. Med. 25, 24–29 (2019).
Hornik, K., Stinchcombe, M. & White, H. Multilayer feedforward networks are universal approximators. Neural Netw. 2, 359–366 (1989).
Wang, M. H. et al. Balancing accuracy and user satisfaction: the role of prompt engineering in AI-driven healthcare solutions. Front. Artif. Intell. 8, 1517918 (2025).
Hayati Rezvan, P., Lee, K. J. & Simpson, J. A. The rise of multiple imputation: a review of the reporting and implementation of the method in medical research. BMC Med. Res. Methodol. 15, 30 (2015).
Jarrett, D., Cebere, B. C., Liu, T., Curth, A. & Schaar, M. van der. HyperImpute: generalized iterative imputation with automatic model selection. In Proc. 39th International Conference on Machine Learning 9916–9937 (PMLR, 2022).
Giuffrè, M. & Shung, D. L. Harnessing the power of synthetic data in healthcare: innovation, application, and privacy. npj Digit.Med. 6, 186 (2023).
Kingma, D. P. & Welling, M. Auto-encoding variational Bayes. Preprint at https://doi.org/10.48550/arXiv.1312.6114 (2013).
Bredell, G., Flouris, K., Chaitanya, K., Erdil, E. & Konukoglu, E. Explicitly minimizing the blur error of variational autoencoders. Preprint at https://doi.org/10.48550/arXiv.2304.05939 (2023).
Goodfellow, I. J. et al. Generative adversarial networks. Preprint at https://doi.org/10.48550/arXiv.1406.2661 (2014).
Arora, A. & Arora, A. Generative adversarial networks and synthetic patient data: current challenges and future perspectives. Future Healthc. J. 9, 190–193 (2022).
Webber, G. & Reader, A. J. Diffusion models for medical image reconstruction. BJRArtificial Intell. 1, ubae013 (2024).
Vivekananthan, S. Comparative analysis of generative models: enhancing image synthesis with VAEs, GANs, and stable diffusion. Preprint at https://doi.org/10.48550/arXiv.2408.08751 (2024).
Khader, F. et al. Denoising diffusion probabilistic models for 3D medical image generation. Sci. Rep. 13, 7303 (2023).
Adams, L. C. et al. What does DALL-E 2 know about radiology? J. Med. Internet Res. 25, e43110 (2023).
Pan, S. et al. Synthetic CT generation from MRI using 3D transformer-based denoising diffusion model. Med. Phys. 51, 2538–2548 (2024).
Inkster, B., Sarda, S. & Subramanian, V. An empathy-driven, conversational artificial intelligence agent (Wysa) for digital mental well-being: real-world data evaluation mixed-methods study. JMIR MHealth UHealth 6, e12106 (2018).
Meinert, E. et al. Accuracy and safety of an autonomous artificial intelligence clinical assistant conducting telemedicine follow-up assessment for cataract surgery. eClinicalMedicine 73, 102692 (2024).
Sackett, C., Harper, D. & Pavez, A. Do we dare use generative AI for mental health?. IEEE Spectr. 61, 42–47 (2024).
Qiu, X. et al. Pre-trained models for natural language processing: A survey. Sci. China E Technol. Sci. 63, 1872–1897 (2020).
Radford, A., Narasimhan, K., Salimans, T. & Sutskever, I. Improving language understanding by generative pre-training. Preprint at https://openai.com/research/language-unsupervised (2018).
Ouyang L, Wu J, Jiang X, et al. Training language models to follow instructions with human feedback. Preprint at https://doi.org/10.48550/arXiv.2203.02155 (2022).
Bai Y, Kadavath S, Kundu S, et al. Constitutional AI: harmlessness from AI feedback. Preprint at https://doi.org/10.48550/arXiv.2212.08073 (2022).
Shao Z, Wang P, Zhu Q, et al. DeepSeekMath: pushing the limits of mathematical reasoning in open language models. Preprint at https://doi.org/10.48550/arXiv.2402.03300 (2024).
Zhou, Y. et al. A foundation model for generalizable disease detection from retinal images. Nature 622, 156–163 (2023).
He, K. et al. Masked autoencoders are scalable vision learners. In Proc. 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 15979–15988 (IEEE, 2022).
Bluethgen, C. et al. A vision–language foundation model for the generation of realistic chest X-ray images. Nat. Biomed. Eng. 9, 494–506 (2024).
Wang, X. et al. A pathology foundation model for cancer diagnosis and prognosis prediction. Nature 634, 970–978 (2024).
Xu, H. et al. A whole-slide foundation model for digital pathology from real-world data. Nature 630, 181–188 (2024).
Pai, S. et al. Foundation model for cancer imaging biomarkers. Nat. Mach. Intell. 6, 354–367 (2024).
Jiao, J. et al. USFM: a universal ultrasound foundation model generalized to tasks and organs towards label efficient image analysis. Med. Image Anal. 96, 103202 (2024).
Qiu, J. et al. Development and validation of a multimodal multitask vision foundation model for generalist ophthalmic artificial intelligence. NEJM AI 1, AIoa2300221 (2024).
Zhou, H.-Y. et al. A transformer-based representation-learning model with unified processing of multimodal input for clinical diagnostics. Nat. Biomed. Eng. 7, 743–755 (2023).
OpenAI. gpt-oss-120b & gpt-oss-20b Model Card. Preprint at https://cdn.openai.com/pdf/419b6906-9da6-406c-a19d-1bb078ac7637/oai_gpt-oss_model_card.pdf (2025).
Llama Team A@ M. Llama 4: leading intelligence. Unrivaled speed and efficiency. Meta https://llama.meta.com/ (2024).
Wei, J. et al. Chain-of-thought prompting elicits reasoning in large language models. In Proc. 36th International Conference on Neural Information Processing Systems 24824–24837 (Curran Associates, 2022).
Hosseini, S. & Seilani, H. The role of agentic AI in shaping a smart future: a systematic review. Array 26, 100399 (2025).
Mukherjee, A. & Chang, H. H. Agentic AI: expanding the algorithmic frontier of creative problem solving. Preprint at https://doi.org/10.48550/arXiv.2502.00289 (2025).
Thirunavukarasu, A. J. et al. Clinical performance of automated machine learning: a systematic review. Ann. Acad. Med. Singap. 53, 187–207 (2024).
Moritz, M., Topol, E. & Rajpurkar, P. Coordinated AI agents for advancing healthcare. Nat. Biomed. Eng. 9, 432–438 (2025).
Tu, T. et al. Towards conversational diagnostic artificial intelligence. Nature 642, 442–450 (2025).
Ananta, I., Khetarpaul, S. & Sharma, D. Symptoms-disease detecting conversation agent using knowledge graphs. In Proc. 2024 Australasian Computer Science Week 98–107 (ACM, 2024).
Alghamdi, H. M. & Mostafa, A. Towards reliable healthcare LLM agents: a case study for pilgrims during Hajj. Information 15, 371 (2024).
Moor, M. et al. Foundation models for generalist medical artificial intelligence. Nature 616, 259–265 (2023).
Magnini, M., Aguzzi, G. & Montagna, S. Open-source small language models for personal medical assistant chatbots. Intell.Based Med. 11, 100197 (2025).
Hinton, G., Vinyals, O. & Dean, J. Distilling the knowledge in a neural network. Preprint at https://doi.org/10.48550/arXiv.1503.02531 (2015).
Muennighoff, N. et al. s1: simple test-time scaling. Preprint at https://doi.org/10.48550/arXiv.2501.19393 (2025).
Kraljevic, Z. et al. Foresight—a generative pretrained transformer for modelling of patient timelines using electronic health records: a retrospective modelling study. Lancet Digit. Health 6, e281–e290 (2024).
Zhang, S. et al. A multimodal biomedical foundation model trained from fifteen million image–text pairs. NEJM AI 2, AIoa2400640 (2025).
Tan, T. F. et al. Artificial intelligence and digital health in global eye health: opportunities and challenges. Lancet Glob. Health 11, e1432–e1443 (2023).
Soltan, A. A. S. et al. A scalable federated learning solution for secondary care using low-cost microcomputing: privacy-preserving development and evaluation of a COVID-19 screening test in UK hospitals. Lancet Digit. Health 6, e93–e104 (2024).
Wahl, B., Cossy-Gantner, A., Germann, S. & Schwalbe, N. R. Artificial intelligence (AI) and global health: how can AI contribute to health in resource-poor settings? BMJ Glob. Health 3, e000798 (2018).
Wang, X. et al. Beyond the limits: a survey of techniques to extend the context length in large language models. Preprint at https://doi.org/10.48550/arXiv.2402.02244 (2024).
Kaplan, J. et al. Scaling laws for neural language models. Preprint at https://doi.org/10.48550/arXiv.2001.08361 (2020).
Yang, R. et al. Retrieval-augmented generation for generative artificial intelligence in health care. npj Health Syst. 2, 1–5 (2025).
Ng, F. Y. C. et al. Artificial intelligence education: an evidence-based medicine approach for consumers, translators, and developers. Cell Rep. Med. 4, 101230 (2023).
Schubert, T., Oosterlinck, T., Stevens, R. D., Maxwell, P. H. & Schaar, M. van der. AI education for clinicians. eClinicalMedicine 79, 102968 (2025).
Shahsavar, Y. & Choudhury, A. User intentions to use ChatGPT for self-diagnosis and health-related purposes: cross-sectional survey study. JMIR Hum. Factors 10, e47564 (2023).
Blease, C. R., Locher, C., Gaab, J., Hägglund, M. & Mandl, K. D. Generative artificial intelligence in primary care: an online survey of UK general practitioners. BMJ Health Care Inform. 31, e101102 (2024).
Gillespie, N., Lockey, S., Ward, T., Macdade, A. & Hassed, G. Trust, attitudes and use of artificial intelligence: a global study 2025. The University of Melbourne and KPMG https://doi.org/10.26188/28822919 (2025).
Jayakumar, S. et al. Quality assessment standards in artificial intelligence diagnostic accuracy systematic reviews: a meta-research study. npj Digit. Med. 5, 11 (2022).
Gilson, A. et al. How does ChatGPT perform on the United States medical licensing examination? The implications of large language models for medical education and knowledge assessment. JMIR Med. Educ. 9, e45312 (2023).
Kung, T. H. et al. Performance of ChatGPT on USMLE:pPotential for AI-assisted medical education using large language models. PLoS Digit. Health 2, e0000198 (2023).
Singhal, K. et al. Large language models encode clinical knowledge. Nature 620, 172–180 (2023).
Thirunavukarasu, A. J. et al. Large language models approach expert-level clinical knowledge and reasoning in ophthalmology: A head-to-head cross-sectional study. PLoS Digit. Health 3, e0000341 (2024).
Ayers, J. W. et al. Comparing physician and artificial intelligence chatbot responses to patient questions posted to a public social media forum. JAMA Intern. Med. 183, 589–596 (2023).
Huo, B. et al. Large language models for chatbot health advice studies: a systematic review. JAMA Netw. Open 8, e2457879 (2025).
Thirunavukarasu, A. J. How can the clinical aptitude of AI assistants be assayed? J. Med. Internet Res. 25, e51603 (2023).
Ebnali Harari, R., Altaweel, A., Ahram, T., Keehner, M. & Shokoohi, H. A randomized controlled trial on evaluating clinician-supervised generative AI for decision support. Int. J. Med. Inf. 195, 105701 (2025).
Xie, Y. et al. Artificial intelligence for teleophthalmology-based diabetic retinopathy screening in a national programme: an economic analysis modelling study. Lancet Digit. Health 2, e240–e249 (2020).
Goh, E. et al. Large language model influence on diagnostic reasoning: a randomized clinical trial. JAMA Netw. Open 7, e2440969 (2024).
Agarwal, N., Moehring, A., Rajpurkar, P. & Salz, T. Combining human expertise with artificial intelligence: experimental evidence from radiology. Working Paper 31422 (NBER, 2023).
Harris, E. Large language models answer medical questions accurately, but can’t match clinicians’ knowledge. JAMA 330, 792–794 (2023).
OpenAI. Reasoning best practices. https://platform.openai.com/docs/guides/reasoning-best-practices (accessed 16 February 2025).
Bedi, S. et al. Testing and evaluation of health care applications of large language models: a systematic review. JAMA 1333, 319–328 (2024).
Kraljevic, Z., Yeung, J. A., Bean, D., Teo, J. & Dobson, R. J. Large language models for medical forecasting – Foresight 2. Preprint at https://doi.org/10.48550/arXiv.2412.10848 (2024).
Kampman, O. P. et al. Conversational self-play for discovering and understanding psychotherapy approaches. Preprint at https://doi.org/10.48550/arXiv.2503.16521 (2025).
Thirunavukarasu, A. J. & O’Logbon, J. The potential and perils of generative artificial intelligence in psychiatry and psychology. Nat. Ment. Health 2, 745–746 (2024).
Siddals, S., Torous, J. & Coxon, A. ‘It happened to be the perfect thing’: experiences of generative AI chatbots for mental health. npj Ment. Health Res. 3, 48 (2024).
Roose, K. Can A.I. be blamed for a teen’s suicide? The New York Times (23 October 2024).
Heaukulani, C., Phang, Y. S., Weng, J. H., Lee, J. & Morris, R. J. T. Deploying AI methods for mental health in Singapore: from mental wellness to serious mental health conditions. Preprint at https://doi.org/10.2139/ssrn.4935469 (2024).
Kampman, O. P. et al. A multi-agent dual dialogue system to support mental health care providers. Preprint at https://doi.org/10.48550/arXiv.2411.18429 (2024).
Brügge, E. et al. Large language models improve clinical decision making of medical students through patient simulation and structured feedback: a randomized controlled trial. BMC Med. Educ. 24, 1391 (2024).
Hale, J., Alexander, S., Wright, S. T. & Gilliland, K. Generative AI in undergraduate medical education: a rapid review. J. Med. Educ. Curric. Dev. 11, 23821205241266697 (2024).
Afzal, S. et al. in Artificial Intelligence in Medicine (eds Michalowski, M. & Moskovitch, R.) 133–145 (Springer International, 2020).
Li, Y. S., Lam, C. S. N. & See, C. Using a machine learning architecture to create an AI-powered chatbot for anatomy education. Med. Sci. Educ. 31, 1729–1730 (2021).
Masters, K. Medical teacher’s first ChatGPT’s referencing hallucinations: lessons for editors, reviewers, and teachers. Med. Teach. 45, 673–675 (2023).
Herd, P. & Moynihan, D. Health care administrative burdens: centering patient experiences. Health Serv. Res. 56, 751–754 (2021).
Wu, D. T. Y. et al. A scoping review of health information technology in clinician burnout. Appl. Clin. Inform. 12, 597–620 (2021).
Coiera, E. & Liu, S. Evidence synthesis, digital scribes, and translational challenges for artificial intelligence in healthcare. Cell Rep. Med. 3, 100860 (2022).
Tierney, A. A. et al. Ambient artificial intelligence scribes to alleviate the burden of clinical documentation. Catal. Non-Issue Content 5, CAT.23.0404 (2024).
Cao, D. Y., Silkey, J. R., Decker, M. C. & Wanat, K. A. Artificial intelligence-driven digital scribes in clinical documentation: pilot study assessing the impact on dermatologist workflow and patient encounters. JAAD Int 15, 149–151 (2024).
Van Veen, D. et al. Adapted large language models can outperform medical experts in clinical text summarization. Nat. Med. https://doi.org/10.1038/s41591-024-02855-5 (2024).
Decker, H. et al. Large language model−based chatbot vs surgeon-generated informed consent documentation for common procedures. JAMA Netw. Open 6, e2336997 (2023).
Gimeno, A., Krause, K., D’Souza, S. & Walsh, C. G. Completeness and readability of GPT-4-generated multilingual discharge instructions in the pediatric emergency department. JAMIA Open 7, ooae050 (2024).
Dong, H. et al. Automated clinical coding: what, why, and where we are?. npj Digit. Med. 5, 1–8 (2022).
Soroush, A. et al. Large language models are poor medical coders — benchmarking of medical code querying. NEJM AI 1, AIdbp2300040 (2024).
Su, X. et al. Multimodal medical code tokenizer. Preprint at https://doi.org/10.48550/arXiv.2502.04397 (2025).
Sanghera, R. et al. High-performance automated abstract screening with large language model ensembles. J. Am. Med. Inform. Assoc. https://doi.org/10.1093/jamia/ocaf050 (2025).
Wornow, M. et al. The shaky foundations of large language models and foundation models for electronic health records. npjDigit. Med. 6, 135 (2023).
Rapp, J. T., Bremer, B. J. & Romero, P. A. Self-driving laboratories to autonomously navigate the protein fitness landscape. Nat. Chem. Eng. 1, 97–107 (2024).
Swanson, K., Wu, W., Bulaong, N. L., Pak, J. E. & Zou, J. The virtual lab of AI agents designs new SARS-CoV-2 nanobodies. Nature https://doi.org/10.1038/s41586-025-09442-9 (2025).
Tayebi Arasteh, S. et al. Large language models streamline automated machine learning for clinical studies. Nat. Commun. 15, 1603 (2024).
Gao, S. et al. Empowering biomedical discovery with AI agents. Cell 187, 6125–6151 (2024).
Suchak, T. et al. Explosion of formulaic research articles, including inappropriate study designs and false discoveries, based on the NHANES US national health database. PLoS Biol. 23, e3003152 (2025).
Alemohammad, S. et al. Self-consuming generative models go MAD. Preprint at https://doi.org/10.48550/arXiv.2307.01850 (2023).
Arora, A. & Arora, A. Synthetic patient data in health care: a widening legal loophole. Lancet 399, 1601–1602 (2022).
Thornton, J. M., Laskowski, R. A. & Borkakoti, N. AlphaFold heralds a data-driven revolution in biology and medicine. Nat. Med. 27, 1666–1669 (2021).
Hayes, T. et al. Simulating 500 million years of evolution with a language model. Science 387, 850–858 (2025).
Nguyen, E. et al. Sequence modeling and design from molecular to genome scale with Evo. Science 386, eado9336 (2024).
Brixi, G. et al. Genome modeling and design across all domains of life with Evo 2. Arc Institute https://arcinstitute.org/manuscripts/Evo2 (accessed 20 February 2025.).
Skarlinski, M. D. et al. Language agents achieve superhuman synthesis of scientific knowledge. Preprint at https://doi.org/10.48550/arXiv.2409.13740 (2024).
Huang, K. et al. Automated hypothesis validation with agentic sequential falsifications. Preprint at https://doi.org/10.48550/arXiv.2502.09858 (2025).
Narayanan, S. et al. Aviary: training language agents on challenging scientific tasks. Preprint at https://doi.org/10.48550/arXiv.2412.21154 (2024).
Gottweis, J. et al. Towards an AI co-scientist. Preprint at https://doi.org/10.48550/arXiv.2502.18864 (2025).
Rajpurkar, P. & Topol, E. J. A clinical certification pathway for generalist medical AI systems. Lancet 405, 20 (2025).
Bedi, S., Shah, N. H. & Koyejo, S. Rethinking evaluation of large language models in healthcare. Competitive Policy International https://www.pymnts.com/cpi-posts/rethinking-evaluation-of-large-language-models-in-healthcare/ (2025).
Yim, D., Khuntia, J., Parameswaran, V. & Meyers, A. Preliminary evidence of the use of generative AI in health care clinical services: systematic narrative review. JMIR Med. Inform. 12, e52073 (2024).
Thirunavukarasu, A. J. et al. Democratizing artificial intelligence imaging analysis with automated machine learning: tutorial. J. Med. Internet Res. 25, e49949 (2023).
Resnik, P. & Lin, J. in The Handbook of Computational Linguistics and Natural Language Processing 271–295 (Wiley Online Library, 2010).
Papineni, K., Roukos, S., Ward, T. & Zhu, W.-J. BLEU: a method for automatic evaluation of machine translation. In Proc. 40th Annual Meeting of the Association for Computational Linguistics (eds. Isabelle, P. et al.) 311–318 (Association for Computational Linguistics, 2002).
Banerjee, S. & Lavie, A. METEOR: an automatic metric for MT evaluation with improved correlation with human judgments. In Proc. ACL Workshop on Intrinsic and Extrinsic Evaluation Measures for Machine Translation and/or Summarization (eds. Goldstein, J. et al.) 65–72 (Association for Computational Linguistics, 2005).
Hossain, E. et al. Natural language processing in electronic health records in relation to healthcare decision-making: a systematic review. Comput. Biol. Med. 155, 106649 (2023).
Haldar, R. & Mukhopadhyay, D. Levenshtein distance technique in dictionary lookup methods: an improved approach. Preprint at https://doi.org/10.48550/arXiv.1101.1232 (2011).
Ganesan, K. ROUGE 2.0: updated and improved measures for evaluation of summarization tasks. Preprint at https://doi.org/10.48550/arXiv.1803.01937 (2018).
Rei, R., Stewart, C., Farinha, A. C. & Lavie, A. COMET: a neural framework for MT evaluation. In Proc. 2020 Conference on Empirical Methods in Natural Language Processing (EMNLP) (eds. Webber, B. et al.) 2685–2702 (Association for Computational Linguistics, 2020).
Tan, T. F. et al. A proposed S.C.O.R.E. evaluation framework for large language models – safety, consensus & context, objectivity, reproducibility and explainability. Preprint at https://doi.org/10.48550/arXiv.2407.07666 (2024).
Zhang, T., Kishore, V., Wu, F., Weinberger, K. Q. & Artzi, Y. BERTScore: evaluating text generation with BERT. Preprint at https://doi.org/10.48550/arXiv.1904.09675 (2019).
Liu, Y. et al. G-Eval: NLG evaluation using GPT-4 with better human alignment. Preprint at https://doi.org/10.48550/arXiv.2303.16634 (2023).
Fu J, Ng SK, Jiang Z, Liu P. GPTScore: evaluate as you desire. Preprint at https://doi.org/10.48550/arXiv.2302.04166 (2023).
Lees, A. et al. A new generation of perspective API: efficient multilingual character-level transformers. In Proc. 28th ACM SIGKDD Conference on Knowledge Discovery and Data Mining 3197–3207 (ACM, 2022).
Teo, C. T. H., Abdollahzadeh, M. & Cheung, N.-M. On measuring fairness in generative models. Preprint at https://doi.org/10.48550/arXiv.2310.19297 (2023).
Min, S. et al. FActScore: fine-grained atomic evaluation of factual precision in long form text generation. Preprint at https://doi.org/10.48550/arXiv.2305.14251 (2023).
Xu, W., Napoles, C., Pavlick, E., Chen, Q. & Callison-Burch, C. Optimizing statistical machine translation for text simplification. Trans. Assoc. Comput. Linguist. 4, 401–415 (2016).
Gu, J. et al. A survey on LLM-as-a-judge. Preprint at https://doi.org/10.48550/arXiv.2411.15594 (2025).
Croxford, E. et al. Automating evaluation of AI text generation in healthcare with a large language model (LLM)-as-a-judge. Preprint at MedRxiv https://doi.org/10.1101/2025.04.22.25326219 (2025).
Abbasian, M. et al. Foundation metrics for evaluating effectiveness of healthcare conversations powered by generative. AI. npj Digit. Med. 7, 82 (2024).
Bommasani, R., Liang, P. & Lee, T. Holistic evaluation of language models. Ann. N Y Acad. Sci. 1525, 140–146 (2023).
Han, R. et al. Randomised controlled trials evaluating artificial intelligence in clinical practice: a scoping review. Lancet Digit. Health 6, e367–e373 (2024).
Gallifant, J. et al. The TRIPOD-LLM reporting guideline for studies using large language models. Nat. Med. 31, 60–69 (2025).
Schulz, K. F., Altman, D. G., Moher, D. & & CONSORT Group CONSORT 2010 statement: updated guidelines for reporting parallel group randomised trials. BMJ 340, c332 (2010).
Huo, B. et al. Reporting guidelines for chatbot health advice studies: explanation and elaboration for the Chatbot Assessment Reporting Tool (CHART). BMJ 390, e083305 (2025).
Chan, A.-W. et al. SPIRIT 2013 statement: defining standard protocol items for clinical trials. Ann. Intern. Med. 158, 200–207 (2013).
Bedi, S. et al. MedHELM: holistic evaluation of large language models for medical tasks. Preprint at https://doi.org/10.48550/arXiv.2505.23802 (2025).
Quin, F., Weyns, D., Galster, M. & Silva, C. C. A/B testing: a systematic literature review. J. Syst. Softw. 211, 112011 (2024).
Austrian, J. et al. Applying A/B testing to clinical decision support: rapid randomized controlled trials. J. Med. Internet Res. 23, e16651 (2021).
Priestman, W. et al. What to expect from electronic patient record system implementation: lessons learned from published evidence. BMJ Health Care Inform. 25, 92–104 (2018).
Ning, Y. et al. Generative artificial intelligence and ethical considerations in health care: a scoping review and ethics checklist. Lancet Digit. Health 6, e848–e856 (2024).
Ning, Y. et al. An ethics assessment tool for artificial intelligence implementation in healthcare: CARE-AI. Nat. Med. 30, 3038–3039 (2024).
Ganapathi, S. et al. Tackling bias in AI health datasets through the STANDING Together initiative. Nat. Med. 28, 2232–2233 (2022).
Khanna, N. N. et al. Economics of artificial intelligence in healthcare: diagnosis vs. treatment. Healthcare 10, 2493 (2022).
Pagallo, U. et al. The underuse of AI in the health sector: opportunity costs, success stories, risks and recommendations. Health Technol. 14, 1–14 (2024).
Nagendran, M. et al. Artificial intelligence versus clinicians: systematic review of design, reporting standards, and claims of deep learning studies. BMJ 368, m689 (2020).
Huo, B. et al. Reporting standards for the use of large language model-linked chatbots for health advice. Nat. Med. 29, 2988 (2023).
Council of the European Union, European Parliament. Regulation (EU) 2024/1689 of the European Parliament and of the Council of 13 June 2024 Laying down Harmonised Rules on Artificial Intelligence and Amending Regulations (EC) No 300/2008, (EU) No 167/2013, (EU) No 168/2013, (EU) 2018/858, (EU) 2018/1139 and (EU) 2019/2144 and Directives 2014/90/EU, (EU) 2016/797 and (EU) 2020/1828 (Artificial Intelligence Act) (Text with EEA Relevance). PE/24/2024/REV/1 (2024).
LCM team et al. Large concept models: language modeling in a sentence representation space. Preprint at https://doi.org/10.48550/arXiv.2412.08821 (2024).
Shen, M., Li, Y., Chen, L. & Yang, Q. From mind to machine: the rise of manus AI as a fully autonomous digital agent. Preprint at https://doi.org/10.48550/arXiv.2505.02024 (2025).
Acknowledgements
National Medical Research Council Singapore (MOH-000655-00/MOH-001014-00), Duke-NUS Medical School (Duke-NUS/RSF/2021/001805/FY2020/EX/15-A5805/FY2022/EX/66-Q128) and Agency for Science, Technology and Research (H20C6a0032).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Medicine thanks Nima Aghaeepour, Madhumita Sushil and Lisa Adams for their contribution to the peer review of this work. Primary Handling Editor: Karen O’Leary, in collaboration with the Nature Medicine team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Teo, Z.L., Thirunavukarasu, A.J., Elangovan, K. et al. Generative artificial intelligence in medicine. Nat Med 31, 3270–3282 (2025). https://doi.org/10.1038/s41591-025-03983-2
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41591-025-03983-2
This article is cited by
-
Artificial intelligence is not a substitute for human intelligence
Nature Reviews Psychology (2025)
-
A novel evaluation benchmark for medical LLMs illuminating safety and effectiveness in clinical domains
npj Digital Medicine (2025)