Fig. 1: Schematic diagram of ColabFold.
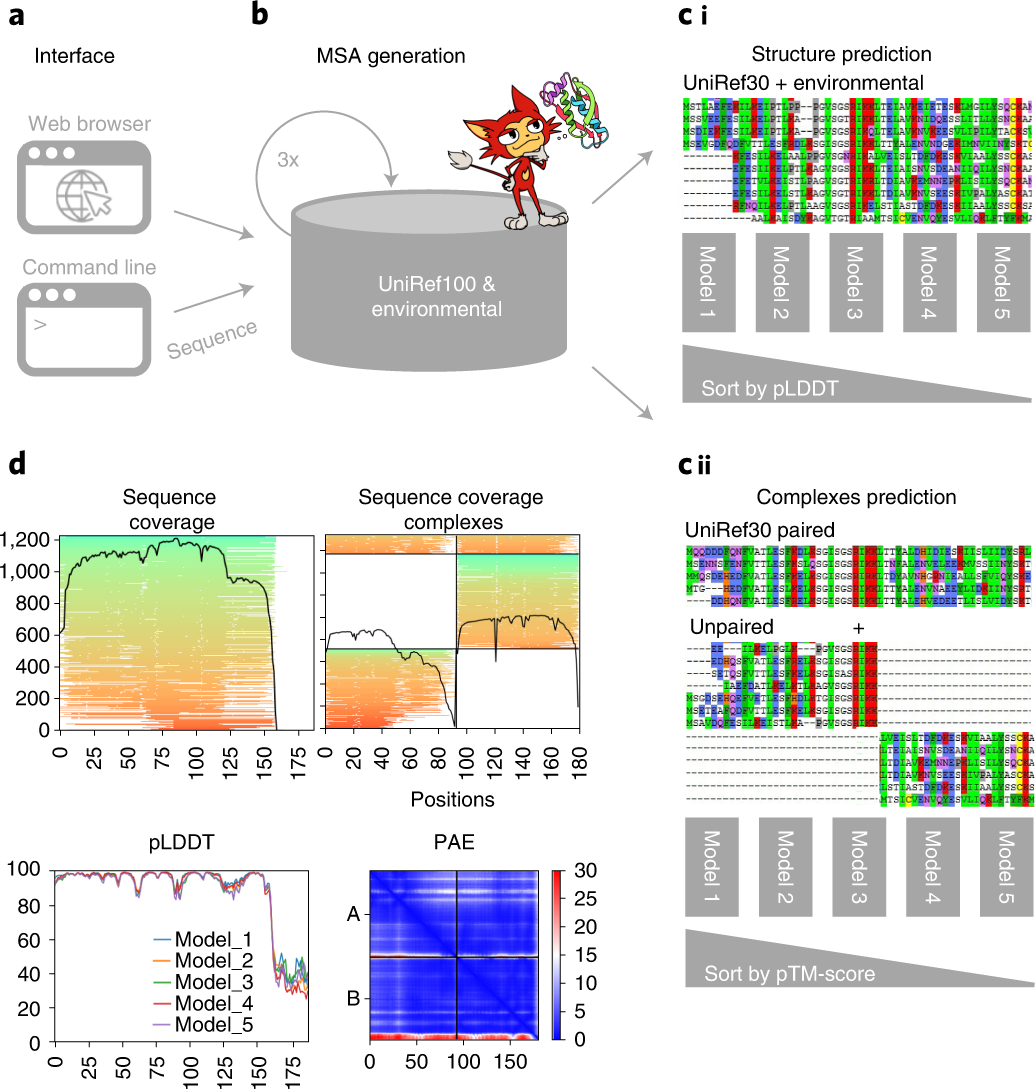
a,b, ColabFold has a web and a command line interface (a) that send FASTA input sequence(s) to an MMseqs2 server (b) searching two databases, UniRef100 and a database of environmental sequences, with three profile-search iterations each. The second database is searched using a sequence profile generated from the UniRef100 search as input. The server generates two MSAs in A3M format containing all detected sequences. c, For predictions of single structures (i) we filter both A3Ms using a diversity-aware filter and return this to be provided as the MSA input feature to the AlphaFold2 models. For predictions of complexes (ii) we pair the top hits within the same species to resolve the inter-chain contacts and additionally add two unpaired MSAs (same as i) to guide the structure prediction. Single chain predictions are ranked by pLDDT and complexes by predicted TM-score. d, To help researchers judge the prediction quality we visualize MSA depth and diversity and show the AlphaFold2 confidence measures (pLDDT and PAE).
