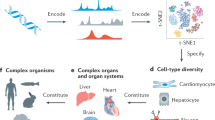
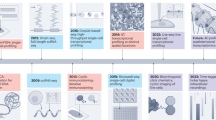
Abstract
To comprehensively understand tissue and organism physiology and pathophysiology, it is essential to create complete three-dimensional (3D) cellular maps. These maps require structural data, such as the 3D configuration and positioning of tissues and cells, and molecular data on the constitution of each cell, spanning from the DNA sequence to protein expression. While single-cell transcriptomics is illuminating the cellular and molecular diversity across species and tissues, the 3D spatial context of these molecular data is often overlooked. Here, I discuss emerging 3D tissue histology techniques that add the missing third spatial dimension to biomedical research. Through innovations in tissue-clearing chemistry, labeling and volumetric imaging that enhance 3D reconstructions and their synergy with molecular techniques, these technologies will provide detailed blueprints of entire organs or organisms at the cellular level. Machine learning, especially deep learning, will be essential for extracting meaningful insights from the vast data. Further development of integrated structural, molecular and computational methods will unlock the full potential of next-generation 3D histology.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Chew, E. J. C. & Tan, P. H. Evolutionary changes in pathology and our understanding of disease. Pathobiology 90, 209–218 (2023).
Tw, H. et al. WormBase: a comprehensive resource for nematode research. Nucleic Acids Res. 38, D463–D467 (2010).
Vergara, H. M. et al. Whole-organism cellular gene-expression atlas reveals conserved cell types in the ventral nerve cord of Platynereis dumerilii. Proc. Natl Acad. Sci. USA 114, 5878–5885 (2017).
Ganesh, K. & Massagué, J. Targeting metastatic cancer. Nat. Med. 27, 34–44 (2021).
Hou, A. J., Chen, L. C. & Chen, Y. Y. Navigating CAR-T cells through the solid-tumour microenvironment. Nat. Rev. Drug Discov. 20, 531–550 (2021).
Raguram, A., Banskota, S. & Liu, D. R. Therapeutic in vivo delivery of gene editing agents. Cell 185, 2806–2827 (2022).
Kimbrel, E. A. & Lanza, R. Next-generation stem cells — ushering in a new era of cell-based therapies. Nat. Rev. Drug Discov. 19, 463–479 (2020).
Tullie, L., Jones, B. C., De Coppi, P. & Li, V. S. W. Building gut from scratch — progress and update of intestinal tissue engineering. Nat. Rev. Gastroenterol. Hepatol. 19, 417–431 (2022).
Brassard, J. A., Nikolaev, M., Hübscher, T., Hofer, M. & Lutolf, M. P. Recapitulating macro-scale tissue self-organization through organoid bioprinting. Nat. Mater. 20, 22–29 (2021).
Tanaka, N. et al. Whole-tissue biopsy phenotyping of three-dimensional tumours reveals patterns of cancer heterogeneity. Nat. Biomed. Eng. 1, 796–806 (2017). The authors demonstrate the power of whole-tissue biopsy phenotyping in revealing patterns of cancer heterogeneity, emphasizing the critical role of 3D characterization in understanding tumor biology.
Xie, W. et al. Prostate cancer risk stratification via nondestructive 3D pathology with deep learning-assisted gland analysis. Cancer Res. 82, 334–345 (2022).
Korteling, J. E., van de Boer-Visschedijk, G. C., Blankendaal, R. A. M., Boonekamp, R. C. & Eikelboom, A. R. Human- versus artificial intelligence. Front. Artif. Intell. 4, 622364 (2021).
Richardson, D. S. et al. Tissue clearing. Nat. Rev. Methods Primers 1, 84 (2021).
Susaki, E. A. & Ueda, H. R. Whole-body and whole-organ clearing and imaging techniques with single-cell resolution: toward organism-level systems biology in mammals. Cell Chem. Biol. 23, 137–157 (2016).
Yang, B. et al. Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell 158, 945–958 (2014).
Belle, M. et al. Tridimensional visualization and analysis of early human development. Cell 169, 161–173 (2017).
Zhao, S. et al. Cellular and molecular probing of intact human organs. Cell 180, 796–812 (2020).
Nectow, A. R. et al. Identification of a brainstem circuit controlling feeding. Cell 170, 429–442 (2017).
Renier, N. et al. Mapping of brain activity by automated volume analysis of immediate early genes. Cell 165, 1789–1802 (2016).
Ren, J. et al. Anatomically defined and functionally distinct dorsal raphe serotonin sub-systems. Cell 175, 472–487 (2018).
Lerner, T. N. et al. Intact-brain analyses reveal distinct information carried by SNc dopamine subcircuits. Cell 162, 635–647 (2015).
Gabanyi, I. et al. Neuro-immune interactions drive tissue programming in intestinal macrophages. Cell 164, 378–391 (2016).
Blain, R. et al. A tridimensional atlas of the developing human head. Cell 186, 5910–5924.e17 (2023).
Murray, E. et al. Simple, scalable proteomic imaging for high-dimensional profiling of intact systems. Cell 163, 1500–1514 (2015).
Chung, K. et al. Structural and molecular interrogation of intact biological systems. Nature 497, 332–337 (2013).
Renier, N. et al. iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell 159, 896–910 (2014).
Mai, H. et al. Whole-body cellular mapping in mouse using standard IgG antibodies. Nat. Biotechnol. 42, 617–627 (2023).
Sylwestrak, E. L., Rajasethupathy, P., Wright, M. A., Jaffe, A. & Deisseroth, K. Multiplexed intact-tissue transcriptional analysis at cellular resolution. Cell 164, 792–804 (2016).
Murakami, T. C. & Heintz, N. Multiplexed and scalable cellular phenotyping toward the standardized three-dimensional human neuroanatomy. Preprint at bioRxiv https://doi.org/10.1101/2022.11.23.517711 (2022).
Kanatani, S. et al. Whole-brain three-dimensional imaging of RNAs at single-cell resolution. Preprint at bioRxiv https://doi.org/10.1101/2022.12.28.521740 (2022).
Zhu, X. et al. Ultrafast optical clearing method for three-dimensional imaging with cellular resolution. Proc. Natl Acad. Sci. USA 116, 11480–11489 (2019).
Kim, S.-Y. et al. Stochastic electrotransport selectively enhances the transport of highly electromobile molecules. Proc. Natl Acad. Sci. USA 112, E6274–E6283 (2015).
Chakraborty, T. et al. Light-sheet microscopy of cleared tissues with isotropic, subcellular resolution. Nat. Methods 16, 1109–1113 (2019).
Gao, L. Extend the field of view of selective plan illumination microscopy by tiling the excitation light sheet. Opt. Express 23, 6102–6111 (2015).
Gao, R. et al. Cortical column and whole-brain imaging with molecular contrast and nanoscale resolution. Science 363, eaau8302 (2019).
Glaser, A. et al. Expansion-assisted selective plane illumination microscopy for nanoscale imaging of centimeter-scale tissues. eLife 12, RP91979 (2023).
Keller, P. J., Schmidt, A. D., Wittbrodt, J. & Stelzer, E. H. K. Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy. Science 322, 1065–1069 (2008).
Chen, Y. et al. A versatile tiling light sheet microscope for imaging of cleared tissues. Cell Rep. 33, 108349 (2020).
Zunino, A. et al. Multiplane encoded light-sheet microscopy for enhanced 3D imaging. ACS Photonics 8, 3385–3393 (2021).
Schueth, A. et al. Efficient 3D light-sheet imaging of very large-scale optically cleared human brain and prostate tissue samples. Commun. Biol. 6, 170 (2023).
Migliori, B. et al. Light sheet theta microscopy for rapid high-resolution imaging of large biological samples. BMC Biol. 16, 57 (2018).
Kim, J. Recent advances in oblique plane microscopy. Nanophotonics 12, 2317–2334 (2023).
Tomer, R., Ye, L., Hsueh, B. & Deisseroth, K. Advanced CLARITY for rapid and high-resolution imaging of intact tissues. Nat. Protoc. 9, 1682–1697 (2014).
Royer, L. A. et al. Adaptive light-sheet microscopy for long-term, high-resolution imaging in living organisms. Nat. Biotechnol. 34, 1267–1278 (2016).
McDole, K. et al. In toto imaging and reconstruction of post-implantation mouse development at the single-cell level. Cell 175, 859–876 (2018).
Matsumoto, K. et al. Advanced CUBIC tissue clearing for whole-organ cell profiling. Nat. Protoc. 14, 3506–3537 (2019).
Liu, T.-L. et al. Observing the cell in its native state: imaging subcellular dynamics in multicellular organisms. Science 360, eaaq1392 (2018).
Silvestri, L. et al. Universal autofocus for quantitative volumetric microscopy of whole mouse brains. Nat. Methods 18, 953–958 (2021).
Conrad, C. et al. Micropilot: automation of fluorescence microscopy-based imaging for systems biology. Nat. Methods 8, 246–249 (2011).
Mahecic, D. et al. Event-driven acquisition for content-enriched microscopy. Nat. Methods 19, 1262–1267 (2022).
Alvelid, J., Damenti, M., Sgattoni, C. & Testa, I. Event-triggered STED imaging. Nat. Methods 19, 1268–1275 (2022).
Shi, Y. et al. Smart lattice light-sheet microscopy for imaging rare and complex cellular events. Nat. Methods 21, 301–310 (2024).
Niehörster, T. et al. Multi-target spectrally resolved fluorescence lifetime imaging microscopy. Nat. Methods 13, 257–262 (2016).
Shi, L. et al. Highly-multiplexed volumetric mapping with Raman dye imaging and tissue clearing. Nat. Biotechnol. 40, 364–373 (2022).
Eng, C.-H. L. et al. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH+. Nature 568, 235–239 (2019).
Chen, K. H., Boettiger, A. N., Moffitt, J. R., Wang, S. & Zhuang, X. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa6090 (2015). Zhuang and colleagues introduce MERFISH, a groundbreaking spatial transcriptomics method that achieves subcellular resolution and features built-in error correction, paving the way for highly multiplexed and precise RNA profiling in single cells.
Ko, J. et al. Spatiotemporal multiplexed immunofluorescence imaging of living cells and tissues with bioorthogonal cycling of fluorescent probes. Nat. Biotechnol. 40, 1654–1662 (2022).
Nichterwitz, S. et al. Laser capture microscopy coupled with Smart-seq2 for precise spatial transcriptomic profiling. Nat. Commun. 7, 12139 (2016).
Mund, A. et al. Deep Visual Proteomics defines single-cell identity and heterogeneity. Nat. Biotechnol. 40, 1231–1240 (2022).
Rosenberger, F. A. et al. Spatial single-cell mass spectrometry defines zonation of the hepatocyte proteome. Nat. Methods 20, 1530–1536 (2023). The study combines microscopy imaging data with ultra-high-sensitivity proteomics, using AI-assisted segmentation, laser microdissection and multiplex-DIA, to achieve deep and interpretable proteomic results at single-cell resolution.
Bhatia, H. S. et al. Spatial proteomics in three-dimensional intact specimens. Cell 185, 5040–5058 (2022). Bhatia et al. present a 3D omics approach that integrates mass spectrometry-based spatial proteomics with cell-level imaging of whole organs and mouse bodies, enabling exploration of the molecular underpinnings of tiny tissue regions identified in large tissues.
Kishi, J. Y. et al. Light-Seq: light-directed in situ barcoding of biomolecules in fixed cells and tissues for spatially indexed sequencing. Nat. Methods 19, 1393–1402 (2022).
Bressan, D., Battistoni, G. & Hannon, G. J. The dawn of spatial omics. Science 381, eabq4964 (2023).
Rodriques, S. G. et al. Slide-seq: a scalable technology for measuring genome-wide expression at high spatial resolution. Science 363, 1463–1467 (2019).
Sudmeier, L. J. et al. Distinct phenotypic states and spatial distribution of CD8+ T cell clonotypes in human brain metastases. Cell Rep. Med. 3, 100620 (2022).
Andersson, A. et al. Spatial deconvolution of HER2-positive breast cancer delineates tumor-associated cell type interactions. Nat. Commun. 12, 6012 (2021).
Moffitt, J. R. et al. Molecular, spatial, and functional single-cell profiling of the hypothalamic preoptic region. Science 362, eaau5324 (2018).
Moncada, R. et al. Integrating microarray-based spatial transcriptomics and single-cell RNA-seq reveals tissue architecture in pancreatic ductal adenocarcinomas. Nat. Biotechnol. 38, 333–342 (2020).
Falconer, D. S., Gauld, I. K. & Roberts, R. C. Cell numbers and cell sizes in organs of mice selected for large and small body size. Genet. Res. 31, 287–301 (1978).
Schede, H. H. et al. Spatial tissue profiling by imaging-free molecular tomography. Nat. Biotechnol. 39, 968–977 (2021).
Angel, T. E. et al. Mass spectrometry based proteomics: existing capabilities and future directions. Chem. Soc. Rev. 41, 3912–3928 (2012).
Rood, J. E. et al. Toward a common coordinate framework for the human body. Cell 179, 1455–1467 (2019).
Wang, Q. et al. The Allen Mouse Brain Common Coordinate Framework: a 3D reference atlas. Cell 181, 936–953 (2020).
Schoppe, O. et al. Deep learning-enabled multi-organ segmentation in whole-body mouse scans. Nat. Commun. 11, 5626 (2020).
Regev, A. et al. The Human Cell Atlas. eLife 6, e27041 (2017).
Heumos, L. et al. Best practices for single-cell analysis across modalities. Nat. Rev. Genet. 24, 550–572 (2023).
Velten, B. & Stegle, O. Principles and challenges of modeling temporal and spatial omics data. Nat. Methods 20, 1462–1474 (2023).
Hallou, A., Yevick, H. G., Dumitrascu, B. & Uhlmann, V. Deep learning for bioimage analysis in developmental biology. Development 148, dev199616 (2021).
Belthangady, C. & Royer, L. A. Applications, promises, and pitfalls of deep learning for fluorescence image reconstruction. Nat. Methods 16, 1215–1225 (2019).
Perakis, A. et al. Contrastive learning of single-cell phenotypic representations for treatment classification. In Machine Learning in Medical Imaging (eds Lian, C. et al.) 565–575 (Springer International, 2021).
Li, X. et al. Real-time denoising enables high-sensitivity fluorescence time-lapse imaging beyond the shot-noise limit. Nat. Biotechnol. 41, 282–292 (2023).
Speiser, A. et al. Deep learning enables fast and dense single-molecule localization with high accuracy. Nat. Methods 18, 1082–1090 (2021).
Weigert, M. et al. Content-aware image restoration: pushing the limits of fluorescence microscopy. Nat. Methods 15, 1090–1097 (2018).
Batson, J. & Royer, L. Noise2Self: blind denoising by self-supervision. Preprint at arxiv.org/abs/1901.11365 (2019).
Jahr, W., Schmid, B., Schmied, C., Fahrbach, F. O. & Huisken, J. Hyperspectral light sheet microscopy. Nat. Commun. 6, 7990 (2015).
Cutrale, F. et al. Hyperspectral phasor analysis enables multiplexed 5D in vivo imaging. Nat. Methods 14, 149–152 (2017).
Krizhevsky, A., Sutskever, I. & Hinton, G. E. ImageNet classification with deep convolutional neural networks. In Advances in Neural Information Processing Systems (eds Pereira, F. et al.) (Curran Associates, 2012). Krizhevsky et al. present a seminal work that demonstrates the power of supervised deep learning for image classification, laying the foundation for the rapid advancements in computer vision and biomedical image analysis.
Chen, J. et al. Three-dimensional residual channel attention networks denoise and sharpen fluorescence microscopy image volumes. Nat. Methods 18, 678–687 (2021).
Haberl, M. G. et al. CDeep3M—plug-and-play cloud-based deep learning for image segmentation. Nat. Methods 15, 677–680 (2018).
Valen, D. A. V. et al. Deep learning automates the quantitative analysis of individual cells in live-cell imaging experiments. PLoS Comput. Biol. 12, e1005177 (2016).
Isensee, F., Jaeger, P. F., Kohl, S. A. A., Petersen, J. & Maier-Hein, K. H. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 18, 203–211 (2021). The authors introduce nnU-Net, a self-configuring variant of the popular U-net architecture that automates key configuration steps for biomedical image segmentation tasks, streamlining the application of deep learning in biomedical image analysis.
Pan, C. et al. Deep learning reveals cancer metastasis and therapeutic antibody targeting in the entire body. Cell 179, 1661–1676 (2019).
Kubota, S. I. et al. Whole-body profiling of cancer metastasis with single-cell resolution. Cell Rep. 20, 236–250 (2017).
Todorov, M. I. et al. Machine learning analysis of whole mouse brain vasculature. Nat. Methods 17, 442–449 (2020).
Kirst, C. et al. Mapping the fine-scale organization and plasticity of the brain vasculature. Cell 180, 780–795 (2020).
Shit, S. et al. clDice — a novel topology-preserving loss function for tubular structure segmentation. In 2021 IEEE/CVF Conference on Computer Vision and Pattern Recognition 16555–16564 (IEEE, 2021).
Stucki, N., Paetzold, J. C., Shit, S., Menze, B. & Bauer, U. Topologically faithful image segmentation via induced matching of persistence barcodes. In Proceedings of the 40th International Conference on Machine Learning (eds Krause, A. et al.) 32698–32727 (PMLR, 2023).
Bai, B. et al. Deep learning-enabled virtual histological staining of biological samples. Light Sci. Appl. 12, 57 (2023).
Christiansen, E. M. et al. In silico labeling: predicting fluorescent labels in unlabeled images. Cell 173, 792–803 (2018).
Rivenson, Y. et al. Virtual histological staining of unlabelled tissue-autofluorescence images via deep learning. Nat. Biomed. Eng. 3, 466–477 (2019). The authors demonstrate virtual histological staining of label-free tissue autofluorescence images using deep learning, potentially replacing time-consuming and costly traditional staining methods
Ghahremani, P. et al. Deep learning-inferred multiplex immunofluorescence for immunohistochemical image quantification. Nat. Mach. Intell. 4, 401–412 (2022).
Ngo, T. B. et al. Label-free cleared tissue microscopy and machine learning for 3D histopathology of biomaterial implants. J. Biomed. Mater. Res. A 111, 840–850 (2023).
Song, A. H. et al. Analysis of 3D pathology samples using weakly supervised AI. Cell 187, 2502–2520.e17 (2024).
Kaltenecker, D. et al. Virtual reality-empowered deep-learning analysis of brain cells. Nat. Methods https://doi.org/10.1038/s41592-024-02245-2 (2024).
Ulman, V. et al. An objective comparison of cell-tracking algorithms. Nat. Methods 14, 1141–1152 (2017).
Sullivan, D. P. et al. Deep learning is combined with massive-scale citizen science to improve large-scale image classification. Nat. Biotechnol. 36, 820–828 (2018). The authors present an innovative approach that harnesses the power of citizen science through computer games to generate large-scale labeled datasets for improving image classification, showcasing the potential of collaborative efforts in addressing the challenge of obtaining high-quality training data for deep learning.
Santos, A. et al. A knowledge graph to interpret clinical proteomics data. Nat. Biotechnol. 40, 692–702 (2022).
Jaume, G. et al. Quantifying explainers of graph neural networks in computational pathology. In Computer Vision and Pattern Recognition Conference 8106–8116 (IEEE, 2021).
Paetzold, J. C. et al. Whole brain vessel graphs: a dataset and benchmark for graph learning and neuroscience. In 35th Conference on Neural Information Processing Systems (eds Ranzato, M. et al.) (NeurIPS, 2021).
Kleshchevnikov, V. et al. Cell2location maps fine-grained cell types in spatial transcriptomics. Nat. Biotechnol. 40, 661–671 (2022).
Dong, R. & Yuan, G.-C. SpatialDWLS: accurate deconvolution of spatial transcriptomic data. Genome Biol. 22, 145 (2021).
Cable, D. M. et al. Robust decomposition of cell type mixtures in spatial transcriptomics. Nat. Biotechnol. 40, 517–526 (2022).
Bergenstråhle, L. et al. Super-resolved spatial transcriptomics by deep data fusion. Nat. Biotechnol. 40, 476–479 (2022).
Zhao, E. et al. Spatial transcriptomics at subspot resolution with BayesSpace. Nat. Biotechnol. 39, 1375–1384 (2021).
Biancalani, T. et al. Deep learning and alignment of spatially resolved single-cell transcriptomes with Tangram. Nat. Methods 18, 1352–1362 (2021).
Hu, J. et al. SpaGCN: integrating gene expression, spatial location and histology to identify spatial domains and spatially variable genes by graph convolutional network. Nat. Methods 18, 1342–1351 (2021).
Jin, L. et al. A comparative study of evaluating missing value imputation methods in label-free proteomics. Sci. Rep. 11, 1760 (2021).
Schoof, E. M. et al. Quantitative single-cell proteomics as a tool to characterize cellular hierarchies. Nat. Commun. 12, 3341 (2021).
Fischer, D. S., Schaar, A. C. & Theis, F. J. Modeling intercellular communication in tissues using spatial graphs of cells. Nat. Biotechnol. 41, 332–336 (2023).
Long, Y. et al. Spatially informed clustering, integration, and deconvolution of spatial transcriptomics with GraphST. Nat. Commun. 14, 1155 (2023). Long et al. present GraphST, a graph-based self-supervised contrastive learning method that outperforms existing tools in spatial clustering, multi-sample integration and cell type deconvolution for spatial transcriptomics data.
Mehrabi, N., Morstatter, F., Saxena, N., Lerman, K. & Galstyan, A. A survey on bias and fairness in machine learning. ACM Comput. Surv. 54, 115 (2021). Mehrabi et al. provide a comprehensive review of researchers’ observations of bias and unfairness in state-of-the-art deep learning, highlighting the critical importance of addressing these issues to ensure the development of equitable and trustworthy AI systems in biomedical research and beyond.
Nauta, M. et al. From anecdotal evidence to quantitative evaluation methods: a systematic review on evaluating explainable AI. ACM Comput. Surv. 55, 295 (2023).
Refaat, A. et al. In vivo fluorescence imaging: success in preclinical imaging paves the way for clinical applications. J. Nanobiotechnology 20, 450 (2022).
Zhang, Q. et al. Adaptive optics for optical microscopy [Invited]. Biomed. Opt. Express 14, 1732–1756 (2023).
Sinitcyn, P. et al. Global detection of human variants and isoforms by deep proteome sequencing. Nat. Biotechnol. 41, 1776–1786 (2023).
BRAIN Initiative Cell Census Network (BICCN). A multimodal cell census and atlas of the mammalian primary motor cortex. Nature 598, 86–102 (2021).
Acknowledgements
This work was supported by the Vascular Dementia Research Foundation, Deutsche Forschungsgemeinschaft (German Research Foundation) under Germany’s Excellence Strategy within the framework of the Munich Cluster for Systems Neurology (EXC 2145 SyNergy, grant 390857198), by a European Research Council Consolidator grant (GA 865323) and a Nomis Heart Atlas Project Grant (Nomis Foundation). I thank M. Elsner and J.C. Paetzold for their scientific input and for editing the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
A.E. is a cofounder of Deep Piction.
Peer review
Peer review information
Nature Methods thanks Christoph Kirst, Ludovico Silvestri, and Raju Tomer for their contribution to the peer review of this work. Primary Handling Editor: Nina Vogt, in collaboration with the Nature Methods team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ertürk, A. Deep 3D histology powered by tissue clearing, omics and AI. Nat Methods 21, 1153–1165 (2024). https://doi.org/10.1038/s41592-024-02327-1
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41592-024-02327-1
This article is cited by
-
Spatial architecture of development and disease
Nature Reviews Genetics (2026)
-
Tissue histology in 3D
Nature Methods (2024)