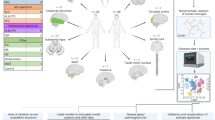
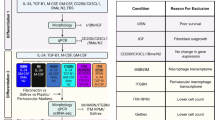
Abstract
The widespread use of single-cell RNA sequencing has generated numerous purportedly distinct and novel subsets of microglia. Here, we challenge this fragmented paradigm by proposing that microglia exist along a continuum rather than as discrete entities. We identify a methodological over-reliance on computational clustering algorithms as the fundamental issue, with arbitrary cluster numbers being interpreted as biological reality. Evidence suggests that the observed transcriptional diversity stems from a combination of microglial plasticity and technical noise, resulting in terminology describing largely overlapping cellular states. We introduce a continuous model of microglial states, where cell positioning along the continuum is determined by biological aging and cell-specific molecular contexts. The model accommodates the dynamic nature of microglia. We advocate for a parsimonious approach toward classification and terminology that acknowledges the continuous spectrum of microglial states, toward a robust framework for understanding these essential immune cells of the CNS.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
The Petilla Interneuron Nomenclature Group (PING). Petilla terminology: nomenclature of features of GABAergic interneurons of the cerebral cortex. Nat. Rev. Neurosci. 9, 557–568 (2008).
Zeng, H. What is a cell type and how to define it? Cell 185, 2739–2755 (2022). This review discusses approaches to the definition of cell types.
Deiters, O. & Schultze, M. J. S. Untersuchungen Über Gehirn Und Rückenmark Des Menschen Und Der Säugethiere (F. Veiweg, 1865).
Golgi, C. Sulla sostanza connettiva del cervello (nevroglia). Rendiconti R. Inst. Lomb. Sci. E Lett. 3, 275–277 (1870).
Zeng, H. & Sanes, J. R. Neuronal cell-type classification: challenges, opportunities and the path forward. Nat. Rev. Neurosci. 18, 530–546 (2017).
BRAIN Initiative Cell Census Network (BICCN). A multimodal cell census and atlas of the mammalian primary motor cortex. Nature 598, 86–102 (2021).
Siletti, K. et al. Transcriptomic diversity of cell types across the adult human brain. Science 382, eadd7046 (2023).
Prinz, M., Masuda, T., Wheeler, M. A. & Quintana, F. J. Microglia and central nervous system-associated macrophages—from origin to disease modulation. Annu. Rev. Immunol. 39, 251–277 (2021).
Amann, L., Masuda, T. & Prinz, M. Mechanisms of myeloid cell entry to the healthy and diseased central nervous system. Nat. Immunol. 24, 393–407 (2023).
Paolicelli, R. C. et al. Microglia states and nomenclature: a field at its crossroads. Neuron 110, 3458–3483 (2022). This is a consensus paper on transcriptional microglia states.
Keren-Shaul, H. et al. A unique microglia type associated with restricting development of Alzheimer’s disease. Cell 169, 1276–1290 (2017). This paper describes AD-associated microglia.
Mathys, H. et al. Single-cell transcriptomic analysis of Alzheimer’s disease. Nature 570, 332–337 (2019).
Masuda, T. et al. Spatial and temporal heterogeneity of mouse and human microglia at single-cell resolution. Nature 566, 388–392 (2019).
Sankowski, R. et al. Mapping microglia states in the human brain through the integration of high-dimensional techniques. Nat. Neurosci. 22, 2098–2110 (2019).
Li, Q. et al. Developmental heterogeneity of microglia and brain myeloid cells revealed by deep single-cell RNA sequencing. Neuron 101, 207–223 (2019).
Safaiyan, S. et al. White matter aging drives microglial diversity. Neuron 109, 1100–1117 (2021).
Tuddenham, J. F. et al. A cross-disease resource of living human microglia identifies disease-enriched subsets and tool compounds recapitulating microglial states. Nat. Neurosci. 27, 2521–2537 (2024).
Chen, Y. & Colonna, M. Microglia in Alzheimer’s disease at single-cell level. Are there common patterns in humans and mice? J. Exp. Med. 218, e20202717 (2021).
Masuda, T., Sankowski, R., Staszewski, O. & Prinz, M. Microglia heterogeneity in the single-cell era. Cell Rep. 30, 1271–1281 (2020).
Böttcher, C. et al. Human microglia regional heterogeneity and phenotypes determined by multiplexed single-cell mass cytometry. Nat. Neurosci. 22, 78–90 (2019).
Olah, M. et al. Single cell RNA sequencing of human microglia uncovers a subset associated with Alzheimer’s disease. Nat. Commun. 11, 6129 (2020).
Venteicher, A. S. et al. Decoupling genetics, lineages, and microenvironment in IDH-mutant gliomas by single-cell RNA-seq. Science 355, eaai8478 (2017).
Darmanis, S. et al. Single-cell RNA-seq analysis of infiltrating neoplastic cells at the migrating front of human glioblastoma. Cell Rep. 21, 1399–1410 (2017).
Müller, S. et al. Single-cell profiling of human gliomas reveals macrophage ontogeny as a basis for regional differences in macrophage activation in the tumor microenvironment. Genome Biol. 18, 234 (2017).
Jordão, M. J. C. et al. Single-cell profiling identifies myeloid cell subsets with distinct fates during neuroinflammation. Science 363, eaat7554 (2019).
Van Hove, H. et al. A single-cell atlas of mouse brain macrophages reveals unique transcriptional identities shaped by ontogeny and tissue environment. Nat. Neurosci. 22, 1021–1035 (2019).
Sankowski, R. et al. Multiomic spatial landscape of innate immune cells at human central nervous system borders. Nat. Med. 30, 186–198 (2024).
Krasemann, S. et al. The TREM2-APOE pathway drives the transcriptional phenotype of dysfunctional microglia in neurodegenerative diseases. Immunity 47, 566–581 (2017). This paper describes AD-associated microglia.
Hammond, T. R. et al. Single-cell RNA sequencing of microglia throughout the mouse lifespan and in the injured brain reveals complex cell-state changes. Immunity 50, 253–271 (2019). This study demonstrates the transcriptional similarity of developmental and aging-associated microglia.
Stadler, T., Pybus, O. G. & Stumpf, M. P. H. Phylodynamics for cell biologists. Science 371, eaah6266 (2021).
Tanay, A. & Sebé-Pedrós, A. Evolutionary cell type mapping with single-cell genomics. Trends Genet. 37, 919–932 (2021).
Arendt, D. et al. The origin and evolution of cell types. Nat. Rev. Genet. 17, 744–757 (2016).
Mrdjen, D. et al. High-dimensional single-cell mapping of central nervous system immune cells reveals distinct myeloid subsets in health, aging, and disease. Immunity 48, 380–395 (2018).
Schulz, C. et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science 336, 86–90 (2012).
Kierdorf, K. et al. Microglia emerge from erythromyeloid precursors via Pu.1- and Irf8-dependent pathways. Nat. Neurosci. 16, 273–280 (2013).
Ginhoux, F. et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 330, 841–845 (2010).
Prinz, M., Jung, S. & Priller, J. Microglia biology: one century of evolving concepts. Cell 179, 292–311 (2019).
Kierdorf, K., Masuda, T., Jordão, M. J. C. & Prinz, M. Macrophages at CNS interfaces: ontogeny and function in health and disease. Nat. Rev. Neurosci. 20, 547–562 (2019).
Masuda, T. et al. Specification of CNS macrophage subsets occurs postnatally in defined niches. Nature 604, 740–748 (2022).
Hickey, W. F. & Kimura, H. Perivascular microglial cells of the CNS are bone marrow-derived and present antigen in vivo. Science 239, 290–292 (1988).
Drieu, A. et al. Parenchymal border macrophages regulate the flow dynamics of the cerebrospinal fluid. Nature 611, 585–593 (2022).
The Tabula Muris Consortium et al. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 562, 367–372 (2018).
The Tabula Muris Consortium et al. A single-cell transcriptomic atlas characterizes ageing tissues in the mouse. Nature 583, 590–595 (2020).
Yao, Z. et al. A high-resolution transcriptomic and spatial atlas of cell types in the whole mouse brain. Nature 624, 317–332 (2023).
Chakarov, S. et al. Two distinct interstitial macrophage populations coexist across tissues in specific subtissular niches. Science 363, eaau0964 (2019).
Hao, Y. et al. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat. Biotechnol. 42, 293–304 (2024).
Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018).
Sankowski, R., Monaco, G. & Prinz, M. Evaluating microglial phenotypes using single-cell technologies. Trends Neurosci. 45, 133–144 (2022).
Tay, T. L. et al. A new fate mapping system reveals context-dependent random or clonal expansion of microglia. Nat. Neurosci. 20, 793–803 (2017).
Réu, P. et al. The lifespan and turnover of microglia in the human brain. Cell Rep. 20, 779–784 (2017).
Kracht, L. et al. Human fetal microglia acquire homeostatic immune-sensing properties early in development. Science 369, 530–537 (2020).
Yousef, H. et al. Aged blood impairs hippocampal neural precursor activity and activates microglia via brain endothelial cell VCAM1. Nat. Med. 25, 988–1000 (2019).
Villeda, S. A. et al. Young blood reverses age-related impairments in cognitive function and synaptic plasticity in mice. Nat. Med. 20, 659–663 (2014).
Hagemeyer, N. et al. Microglia contribute to normal myelinogenesis and to oligodendrocyte progenitor maintenance during adulthood. Acta Neuropathol. 134, 441–458 (2017).
Lan, Y. et al. Fate mapping of Spp1 expression reveals age-dependent plasticity of disease-associated microglia-like cells after brain injury. Immunity 57, 349–363 (2024).
Barclay, K. M. et al. An inducible genetic tool to track and manipulate specific microglial states reveals their plasticity and roles in remyelination. Immunity 57, 1394–1412 (2024).
De Vlaminck, K. et al. Differential plasticity and fate of brain-resident and recruited macrophages during the onset and resolution of neuroinflammation. Immunity 55, 2085–2102 (2022).
Stratoulias, V. et al. ARG1-expressing microglia show a distinct molecular signature and modulate postnatal development and function of the mouse brain. Nat. Neurosci. 26, 1008–1020 (2023).
Escoubas, C. C. et al. Type-I-interferon-responsive microglia shape cortical development and behavior. Cell 187, 1936–1954 (2024).
Datta, M. et al. Histone deacetylases 1 and 2 regulate microglia function during development, homeostasis, and neurodegeneration in a context-dependent manner. Immunity 48, 514–529 (2018).
Zierfuss, B. et al. Targeting foam cell formation in inflammatory brain diseases by the histone modifier MS‐275. Ann. Clin. Transl. Neurol. 7, 2161–2177 (2020).
Hoeksema, M. A. et al. Targeting macrophage histone deacetylase 3 stabilizes atherosclerotic lesions. EMBO Mol. Med. 6, 1124–1132 (2014). This study shows the involvement of histone deacetylases in phagocyte polarization, suggesting a molecular point-of-no-return.
Hayflick, L. & Moorhead, P. S. The serial cultivation of human diploid cell strains. Exp. Cell. Res. 25, 585–621 (1961). This study proposes an upper limit for the number of replication cycles for normal cells, known as the Hayflick limit.
Hu, Y. et al. Replicative senescence dictates the emergence of disease-associated microglia and contributes to Aβ pathology. Cell Rep. 35, 109228 (2021).
Rachmian, N. et al. Identification of senescent, TREM2-expressing microglia in aging and Alzheimer’s disease model mouse brain. Nat. Neurosci. 27, 1116–1124 (2024).
Li, X. et al. Transcriptional and epigenetic decoding of the microglial aging process. Nat. Aging 3, 1288–1311 (2023).
Wendeln, A. -C. et al. Innate immune memory in the brain shapes neurological disease hallmarks. Nature 556, 332–338 (2018).
Kinker, G. S. et al. Pan-cancer single-cell RNA-seq identifies recurring programs of cellular heterogeneity. Nat. Genet. 52, 1208–1218 (2020). This study shows that tumor cells are present in recurrent states across different tumor entities.
Neftel, C. et al. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell 178, 835–849 (2019). This paper demonstrates the fluidity of glioblastoma transcriptional states.
Sun, N. et al. Human microglial state dynamics in Alzheimer’s disease progression. Cell 186, 4386–4403 (2023).
Mancuso, R. et al. Xenografted human microglia display diverse transcriptomic states in response to Alzheimer's disease-related amyloid-β pathology. Nat. Neurosci. 27, 886–900 (2024).
Sala Frigerio, C. et al. The major risk factors for Alzheimer’s disease: age, sex, and genes modulate the microglia response to Aβ plaques. Cell Rep. 27, 1293–1306 (2019).
Shukla, A. K. et al. CD11a expression distinguishes infiltrating myeloid cells from plaque‐associated microglia in Alzheimer’s disease. Glia 67, 844–856 (2019).
Yin, Z. et al. Immune hyperreactivity of Aβ plaque-associated microglia in Alzheimer’s disease. Neurobiol. Aging 55, 115–122 (2017).
Grubman, A. et al. Transcriptional signature in microglia associated with Aβ plaque phagocytosis. Nat. Commun. 12, 3015 (2021).
Marschallinger, J. et al. Lipid-droplet-accumulating microglia represent a dysfunctional and proinflammatory state in the aging brain. Nat. Neurosci. 23, 194–208 (2020).
Gazestani, V. et al. Early Alzheimer’s disease pathology in human cortex involves transient cell states. Cell 186, 4438–4453 (2023).
Bakken, T. E. et al. Comparative cellular analysis of motor cortex in human, marmoset and mouse. Nature 598, 111–119 (2021).
Wang, Z. et al. An immune cell atlas reveals the dynamics of human macrophage specification during prenatal development. Cell 186, 4454–4471 (2023).
McKenna, A. et al. Whole-organism lineage tracing by combinatorial and cumulative genome editing. Science 353, aaf7907 (2016).
Lareau, C. A. et al. Massively parallel single-cell mitochondrial DNA genotyping and chromatin profiling. Nat. Biotechnol. 39, 451–461 (2021). This paper demonstrates an approach to infer cell clonality from mitochondrial single-nucleotide variants.
Pei, W. et al. Polylox barcoding reveals haematopoietic stem cell fates realized in vivo. Nature 548, 456–460 (2017).
Trapnell, C. Defining cell types and states with single-cell genomics. Genome Res. 25, 1491–1498 (2015).
Morris, S. A. The evolving concept of cell identity in the single cell era. Development 146, dev169748 (2019).
Crowell, H. L. et al. muscat detects subpopulation-specific state transitions from multi-sample multi-condition single-cell transcriptomics data. Nat. Commun. 11, 6077 (2020).
Grün, D. Revealing dynamics of gene expression variability in cell state space. Nat. Methods 17, 45–49 (2020).
Karczewski, K. J. & Snyder, M. P. Integrative omics for health and disease. Nat. Rev. Genet. 19, 299–310 (2018).
Clarke, R. et al. The properties of high-dimensional data spaces: implications for exploring gene and protein expression data. Nat. Rev. Cancer 8, 37–49 (2008).
Hotelling, H. Analysis of a complex of statistical variables into principal components. J. Educ. Psychol. 24, 417–441 (1933).
Wang, A. & Gehan, E. A. Gene selection for microarray data analysis using principal component analysis. Stat. Med. 24, 2069–2087 (2005).
Maaten van der, L. & Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 (2008).
McInnes, L., Healy, J. & Melville, J. UMAP: uniform manifold approximation and projection for dimension reduction. Preprint at https://doi.org/10.48550/arXiv.1802.03426 (2018).
Silverman, J. D., Roche, K., Mukherjee, S. & David, L. A. Naught all zeros in sequence count data are the same. Comput. Struct. Biotechnol. J. 18, 2789–2798 (2020).
Kharchenko, P. V., Silberstein, L. & Scadden, D. T. Bayesian approach to single-cell differential expression analysis. Nat. Methods 11, 740–742 (2014).
Grün, D., Kester, L. & van Oudenaarden, A. Validation of noise models for single-cell transcriptomics. Nat. Methods 11, 637–640 (2014).
Haque, A., Engel, J., Teichmann, S. A. & Lönnberg, T. A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications. Genome Med. 9, 75 (2017).
Luecken, M. D. & Theis, F. J. Current best practices in single‐cell RNA‐seq analysis: a tutorial. Mol. Syst. Biol. 15, e8746 (2019).
Tjärnberg, A. et al. Optimal tuning of weighted kNN- and diffusion-based methods for denoising single cell genomics data. PLoS Comput. Biol. 17, e1008569 (2021).
Stuart, T. & Satija, R. Integrative single-cell analysis. Nat. Rev. Genet. 20, 257–272 (2019).
Welch, J. et al. Integrative inference of brain cell similarities and differences from single-cell genomics. Preprint at bioRxiv https://doi.org/10.1101/459891 (2018).
Acknowledgements
R.S. is supported by the IMMediate Advanced Clinician Scientist-Program, Department of Medicine II, Medical Center, University of Freiburg and Faculty of Medicine, University of Freiburg, funded by the Bundesministerium für Bildung und Forschung (Federal Ministry of Education and Research), 01EO2103. Furthermore, R.S. is supported by the Fritz Thyssen Foundation. M.P. is supported by the DFG (CRC/TRR167 project ID 259373024 ‘NeuroMac’). M.P. is further supported by the Sobek Foundation, the Ernst-Jung Foundation, the Klaus Faber Foundation, the Novo Nordisk Prize, the German Research Foundation (SFB 992 project ID 192904750, SFB 1160, SFB-1479 project ID 441891347, TRR 359 project ID 491676693 and Gottfried Wilhelm Leibniz Prize) and by the DFG under Germany’s Excellence Strategy (CIBSS EXC-2189 project ID 390939984). The funders had no role in the study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Neuroscience thanks Marta Olah and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sankowski, R., Prinz, M. A dynamic and multimodal framework to define microglial states. Nat Neurosci 28, 1372–1380 (2025). https://doi.org/10.1038/s41593-025-01978-3
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41593-025-01978-3
This article is cited by
-
Making tracks: microglia and the extracellular matrix
Molecular Neurodegeneration (2025)