Figure 1
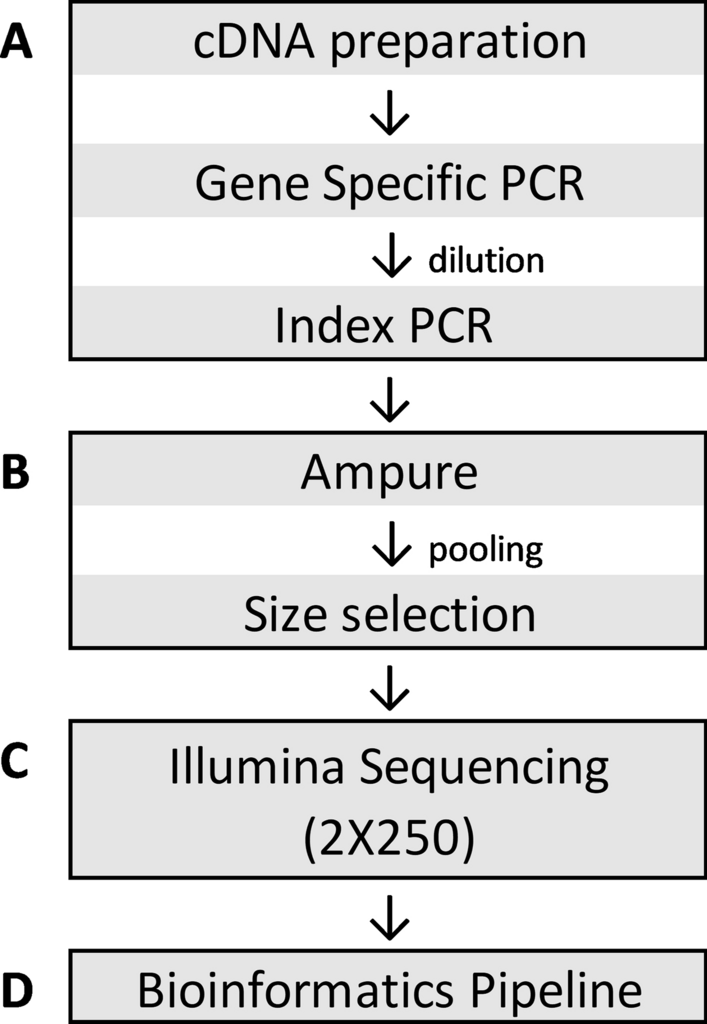
High-throughput NGS workflow. Sequencing library preparation was performed on robotic liquid handlers. (A) Extracted RNA of SARS-CoV-2 positive specimens were converted to cDNA and amplified using a touchdown PCR method with primers published by the ARTIC network with modifications to add Illumina sequencing primer binding sites. Specimen-specific indexed sequencing adapters were added by subsequent fusion PCR using the primer binding sites. Agilent Bravo was used for each process. (B) The final indexed PCR products were purified using Ampure XP beads using BlueCat BlueWasher, pooled into a single library using Hamilton Starlet, and size selected using Sage Science Blue Pippin. (C) The final library was sequenced on an Illumina NovaSeq 6000. (D) An in-house developed bioinformatics pipeline was utilized to generate consensus genomes and for variant calls relative to the reference genome MN908947.3, Wuhan-Hu-1.
