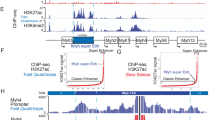
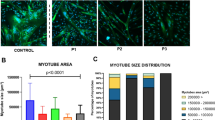
Abstract
Skeletal muscles are essential for movement, supporting a wide range of locomotor behaviors. Muscle tissue is composed of multiple cell types including “fast” and “slow” myofibers, whose contractile properties are largely influenced by selective expression of myosin heavy chain (MyHC) isoforms. While ‘super-enhancers’ regulating MyHC gene clusters have been identified, the cis-regulatory elements (CREs) controlling non-MyHC genes important to myofiber physiology remain less defined. Here, we profile the regulatory landscape of two pairs of mouse hind limb muscles differing in MyHC expression at a late embryonic (E18.5) and adult time point to identify candidate CREs that may regulate genes important to myofiber type. Gene expression and chromatin accessibility analyses revealed that epigenetic differences at E18.5 largely reflect limb patterning, whereas adult differences reflect myofiber differentiation. We identified thousands of differentially accessible regions that may regulate genes important for muscle development, muscle biology, and myofiber identity. Among these, twelve conserved, muscle-specific CREs associated with myofiber type were tested for regulatory activity. Nine enhanced and three reduced gene activity in vitro, although their phenotypic effects remain unknown. By profiling multiple muscles across two time points, our study extends current understanding of conserved, muscle-specific CREs that regulate gene expression during myogenesis.
Similar content being viewed by others
Data availability
All ATAC-seq and RNA-seq datasets generated as part of this study have been deposited to the NCBI GEO repository under accession number GSE292230 and GSE292232. All data included in this study are available upon reasonable request by contact with the corresponding author.
References
Janssen, I., Heymsfield, S. B., Wang, Z. & Ross, R. Skeletal muscle mass and distribution in 468 men and women aged 18–88 year. J. Appl. Physiol. 89, 81–88 (2000).
Zihlman, A. L. & Bolter, D. R. Body composition in Pan Paniscus compared with homo sapiens has implications for changes during human evolution. Proc. Natl. Acad. Sci. 112, 7466–7471 (2015).
O’Neill, M. C., Umberger, B. R., Holowka, N. B., Larson, S. G. & Reiser, P. J. Chimpanzee super strength and human skeletal muscle evolution. In: Proceedings of the National Academy of Sciences. 114 7343–7348 (2017).
King, A. M., Loiselle, D. S. & Kohl, P. Force generation for locomotion of vertebrates: skeletal muscle overview. IEEE J. Oceanic Eng. 29, 684–691 (2004).
Brooks, S. V. Current topics for teaching skeletal muscle physiology. Adv. Physiol. Educ. 27, 171–182 (2003).
Periasamy, M., Herrera, J. L. & Reis, F. C. G. Skeletal muscle thermogenesis and its role in whole body energy metabolism. Diabetes Metab. J. 41, 327 (2017).
Bourey, R. E., Koranyi, L., James, D. E., Mueckler, M. & Permutt, M. A. Effects of altered glucose homeostasis on glucose transporter expression in skeletal muscle of the rat. J. Clin. Invest. 86, 542–547 (1990).
Wolfe, R. R. The underappreciated role of muscle in health and disease. Am. J. Clin. Nutr. 84, 475–482 (2006).
Moreno-Justicia, R. et al. Human skeletal muscle fiber heterogeneity beyond myosin heavy chains. Nat. Commun. 16, 1764 (2025).
Murgia, M. et al. Protein profile of fiber types in human skeletal muscle: a single-fiber proteomics study. Skelet. Muscle. 11, 24 (2021).
Stuart, C. A. et al. Myosin content of individual human muscle fibers isolated by laser capture microdissection. Am. J. Physiology-Cell Physiol. 310, C381–C389 (2016).
Petrany, M. J. et al. Single-nucleus RNA-seq identifies transcriptional heterogeneity in multinucleated skeletal myofibers. Nat. Commun. 11, 6374 (2020).
Howald, H., Hoppeler, H., Claassen, H., Mathieu, O. & Straub, R. Influences of endurance training on the ultrastructural composition of the different muscle fiber types in humans. Pflügers Archive Eur. J. Physiol. 403, 369–376 (1985).
Bárány, M. ATPase activity of myosin correlated with speed of muscle shortening. J. Gen. Physiol. 50, 197–218 (1967).
Schiaffino, S. & Reggiani, C. Fiber types in mammalian skeletal muscles. Physiol. Rev. 91, 1447–1531 (2011).
Schiaffino, S., Reggiani, C., Kostrominova, T. Y., Mann, M. & Murgia, M. Mitochondrial specialization revealed by single muscle fiber proteomics: focus on the Krebs cycle. Scand. J. Med. Sci. Sports. 25, 41–48 (2015).
Edman, S., Flockhart, M., Larsen, F. J. & Apró, W. Need for speed: human fast-twitch mitochondria favor power over efficiency. Mol. Metab. 79, 101854 (2024).
Schiaffino, S., Rossi, A. C., Smerdu, V., Leinwand, L. A. & Reggiani, C. Developmental myosins: expression patterns and functional significance. Skelet. Muscle. 5, 22 (2015).
Pette, D. & Staron, R. S. Myosin isoforms, muscle fiber types, and transitions. Microsc Res. Tech. 50, 500–509 (2000).
Burke, R. E., Levine, D. N., Zajac, F. E., Tsairis, P. & Engel, W. K. Mammalian motor units: Physiological-histochemical correlation in three types in Cat gastrocnemius. Sci. (1979). 174, 709–712 (1971).
Bottinelli, R. & Reggiani, C. Human skeletal muscle fibres: molecular and functional diversity. Prog Biophys. Mol. Biol. 73, 195–262 (2000).
Resnicow, D. I., Deacon, J. C., Warrick, H. M., Spudich, J. A. & Leinwand, L. A. Functional diversity among a family of human skeletal muscle myosin motors. In: Proceedings of the National Academy of Sciences. 107 1053–1058 (2010).
Hagiwara, N., Yeh, M. & Liu, A. Sox6 is required for normal fiber type differentiation of fetal skeletal muscle in mice. Dev. Dyn. 236, 2062–2076 (2007).
Hennebry, A. et al. Myostatin regulates fiber-type composition of skeletal muscle by regulating MEF2 and myod gene expression. Am. J. Physiology-Cell Physiol. 296, C525–C534 (2009).
Chin, E. R. et al. A calcineurin-dependent transcriptional pathway controls skeletal muscle fiber type. Genes Dev. 12, 2499–2509 (1998).
Buller, A. J., Eccles, J. C. & Eccles, R. M. Differentiation of fast and slow muscles in the Cat Hind limb. J. Physiol. 150, 399–416 (1960).
Agbulut, O., Noirez, P. & Beaumont, F. Butler-Browne, G. Myosin heavy chain isoforms in postnatal muscle development of mice. Biol. Cell. 95, 399–406 (2003).
Arakelian, C. et al. Myosin S2 origins track evolution of strong binding on actin by azimuthal rolling of motor domain. Biophys. J. 108, 1495–1502 (2015).
Mascarello, F., Toniolo, L., Cancellara, P., Reggiani, C. & Maccatrozzo, L. Expression and identification of 10 sarcomeric MyHC isoforms in human skeletal muscles of different embryological origin. Diversity and similarity in mammalian species. Annals Anat. - Anatomischer Anzeiger. 207, 9–20 (2016).
Mishra, P., Varuzhanyan, G., Pham, A. H. & Chan, D. C. Mitochondrial dynamics is a distinguishing feature of skeletal muscle fiber types and regulates organellar compartmentalization. Cell. Metab. 22, 1033–1044 (2015).
Callahan, D. M., Umberger, B. R. & Kent, J. A. Mechanisms of in vivo muscle fatigue in humans: investigating age-related fatigue resistance with a computational model. J. Physiol. 594, 3407–3421 (2016).
Cooke, R., Franks, K., Luciani, G. B. & Pate, E. The Inhibition of rabbit skeletal muscle contraction by hydrogen ions and phosphate. J. Physiol. 395, 77–97 (1988).
Debold, E. P., Dave, H. & Fitts, R. H. Fiber type and temperature dependence of inorganic phosphate: implications for fatigue. Am. J. Physiology-Cell Physiol. 287, C673–C681 (2004).
Debold, E. P., Beck, S. E. & Warshaw, D. M. Effect of low pH on single skeletal muscle myosin mechanics and kinetics. Am. J. Physiology-Cell Physiol. 295, C173–C179 (2008).
Barclay, C. J., Constable, J. K. & Gibbs, C. L. Energetics of fast- and slow‐twitch muscles of the mouse. J. Physiol. 472, 61–80 (1993).
Barclay, C. J. Efficiency of Fast- and Slow-Twitch muscles of the mouse performing Cyclic contractions. J. Exp. Biol. 193, 65–78 (1994).
Queeno, S. R. et al. Human and African ape myosin heavy chain content and the evolution of hominin skeletal muscle. Comp. Biochem. Physiol. Mol. Integr. Physiol. 281, 111415 (2023).
Spainhower, K. B. et al. Coming to grips with life upside down: how myosin fiber type and metabolic properties of sloth hindlimb muscles contribute to suspensory function. J. Comp. Physiol. B. 191, 207–224 (2021).
Kimura, T., Kumakura, H., Inokuchi, S. & Ishida, H. Composition of muscle fibers in the slow loris, using the m. biceps brachii as an example. Primates 28, 525–532 (1987).
Okerblom, J. et al. Human-like Cmah inactivation in mice increases running endurance and decreases muscle fatigability: implications for human evolution. Proc. Royal Soc. B: Biol. Sci. 285, 20181656 (2018).
Poole, D. C. & Erickson, H. H. Highly Athletic Terrestrial Mammals: Horses and Dogs. In Comprehensive Physiology (Wiley, 2011). https://doi.org/10.1002/cphy.c091001.
LaPotin, S. et al. Divergent cis-regulatory evolution underlies the convergent loss of sodium channel expression in electric fish. Sci Adv 8, https://doi.org/10.1126/sciadv.abm2970 (2022).
Röckel, F. et al. Color intensity of the Red-Fleshed berry phenotype of vitis vinifera teinturier grapes varies due to a 408 bp duplication in the promoter of VvmybA1. Genes (Basel). 11, 891 (2020).
Aldea, D. et al. Repeated mutation of a developmental enhancer contributed to human thermoregulatory evolution. In: Proceedings of the National Academy of Sciences. 118 (2021).
Gokhman, D. et al. Differential DNA methylation of vocal and facial anatomy genes in modern humans. Nat. Commun. 11, 1189 (2020).
Kvon, E. Z. et al. Progressive loss of function in a limb enhancer during snake evolution. Cell 167, 633–642e11 (2016).
Sicard, A. et al. Standing genetic variation in a tissue-specific enhancer underlies selfing-syndrome evolution in capsella. Proc. Natl. Acad. Sci. 113, 13911–13916 (2016).
Wang, X. et al. Genetic variation in ZmVPP1 contributes to drought tolerance in maize seedlings. Nat. Genet. 48, 1233–1241 (2016).
Jiang, P. & Rausher, M. Two genetic changes in cis-regulatory elements caused evolution of petal spot position in Clarkia. Nat. Plants. 4, 14–22 (2018).
Kratochwil, C. F. et al. Agouti-related peptide 2 facilitates convergent evolution of Stripe patterns across cichlid fish radiations. Sci. (1979). 362, 457–460 (2018).
Letelier, J. et al. A conserved Shh cis-regulatory module highlights a common developmental origin of unpaired and paired fins. Nat. Genet. 50, 504–509 (2018).
Roeske, M. J., Camino, E. M., Grover, S., Rebeiz, M. & Williams, T. M. Cis-regulatory evolution integrated the Bric-à-brac transcription factors into a novel fruit fly gene regulatory network. Elife 7, e32273 (2018).
Thompson, A. C. et al. A novel enhancer near the Pitx1 gene influences development and evolution of pelvic appendages in vertebrates. Elife 7, e38555 (2018).
Lewis, J. J. et al. Parallel evolution of ancient, pleiotropic enhancers underlies butterfly wing pattern mimicry. In: Proceedings of the National Academy of Sciences. 116 24174–24183 (2019).
Dos Santos, M. et al. A fast myosin super enhancer dictates muscle fiber phenotype through competitive interactions with myosin genes. Nat. Commun. 13, 1039 (2022).
Long, K. et al. Identification of enhancers responsible for the coordinated expression of myosin heavy chain isoforms in skeletal muscle. BMC Genom. 23, 519 (2022).
Ramachandran, K. et al. Dynamic enhancers control skeletal muscle identity and reprogramming. PLoS Biol. 17, e3000467 (2019).
Belotti, E. et al. H2A.Z is dispensable for both basal and activated transcription in post-mitotic mouse muscles. Nucleic Acids Res. 48, 4601–4613 (2020).
Dos Santos, M. et al. Single-nucleus RNA-seq and FISH identify coordinated transcriptional activity in mammalian myofibers. Nat. Commun. 11, 5102 (2020).
Rovito, D. et al. Myod1 and GR coordinate myofiber-specific transcriptional enhancers. Nucleic Acids Res. 49, 4472–4492 (2021).
Sahinyan, K. et al. Application of ATAC-Seq for genome-wide analysis of the chromatin state at single myofiber resolution. Elife 11, e72792 (2022).
Lin, H. et al. Reprogramming of cis-regulatory networks during skeletal muscle atrophy in male mice. Nat. Commun. 14, 6581 (2023).
Blackburn, D. M. et al. The E3 ubiquitin ligase Nedd4L preserves skeletal muscle stem cell quiescence by inhibiting their activation. iScience 27, 110241 (2024).
Garcia, P. et al. Setdb1 protects genome integrity in murine muscle stem cells to allow for regenerative myogenesis and inflammation. Dev. Cell. 59, 2375–2392e8 (2024).
Dos Santos, M. et al. Opposing gene regulatory programs governing myofiber development and maturation revealed at single nucleus resolution. Nat. Commun. 14, 4333 (2023).
National Research Council (U.S.). Committee for the Update of the Guide for the Care and Use of Laboratory Animals., Institute for Laboratory Animal Research (U.S.) & National Academies Press (U.S.). Guide for the Care and Use of Laboratory Animals. https://doi.org/10.17226/12910 (National Academies Press, 2011).
Kilkenny, C., Browne, W., Cuthill, I. C., Emerson, M. & Altman, D. G. Animal research: reporting in vivo experiments: the ARRIVE guidelines. Br. J. Pharmacol. 160, 1577–1579 (2010).
Wigmore, P. M. & Dunglison, G. F. The generation of fiber diversity during myogenesis. Int. J. Dev. Biol. 42, 117–125 (1998).
Staack, A., Donjacour, A. A., Brody, J., Cunha, G. R. & Carroll, P. Mouse urogenital development: a practical approach. Differentiation 71, 402–413 (2003).
Terry, E. E. et al. Transcriptional profiling reveals extraordinary diversity among skeletal muscle tissues. Elife 7, e34613 (2018).
Burkholder, T. J., Fingado, B., Baron, S. & Lieber, R. L. Relationship between muscle fiber types and sizes and muscle architectural properties in the mouse hindlimb. J. Morphol. 221, 177–190 (1994).
Asmussen, G. & Gaunitz, U. Temperature effects on isometric contractions of slow and fast twitch muscles of various rodents–dependence on fibre type composition: a comparative study. Biomed. Biochim. Acta. 48, S536–S541 (1989).
Augusto, V., Padovani, C. R. & Rocha Campos, G. E. Skeletal muscle fiber types in C57Bl6J mice. Brazilian J. Morphological Sci. 21, 89–94 (2004).
Bloemberg, D. & Quadrilatero, J. Rapid determination of myosin heavy chain expression in rat, mouse, and human skeletal muscle using multicolor immunofluorescence analysis. PLoS One https://doi.org/10.1371/journal.pone.0035273 (2012).
Hämäläinen, N. & Pette, D. The histochemical profiles of fast fiber types IIB, IID, and IIA in skeletal muscles of mouse, rat, and rabbit. J. Histochem. Cytochemistry. 41, 733–743 (1993).
Hitomi, Y. et al. Seven skeletal muscles rich in slow muscle fibers May function to sustain neutral position in the rodent hindlimb. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 140, 45–50 (2005).
Minchew, E. C., Williamson, N. C., Readyoff, A. T., McClung, J. M. & Spangenburg, E. E. Isometric skeletal muscle contractile properties in common strains of male laboratory mice. Front. Physiol. https://doi.org/10.3389/fphys.2022.937132 (2022).
Brasseur, J. E. et al. Systematic distribution of muscle fiber types in the medical gastrocnemius of the laboratory mouse: A morphometric analysis. Anat. Rec. 218, 396–401 (1987).
Charles, J. P., Cappellari, O., Spence, A. J., Hutchinson, J. R. & Wells, D. J. Musculoskeletal Geometry, muscle architecture and functional specialisations of the mouse hindlimb. PLoS One. 11, e0147669 (2016).
Hitz, B. C. et al. The ENCODE uniform analysis pipelines. bioRxiv https://doi.org/10.1101/2023.04.04.535623 (2023).
Racine, J. S. & RStudio: A Platform-Independent IDE for R and Sweave. J. Appl. Econom. 27, 167–172 (2012).
R Core Team. R: A Language and Environment for Statistical Computing. Preprint at https://www.R-project.org/ (2024).
Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Preprint at http://www.bioinformatics.babraham.ac.uk/projects/fastqc (2015).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Deng, Z. L., Münch, P. C., Mreches, R. & McHardy, A. C. Rapid and accurate identification of ribosomal RNA sequences via deep learning. Nucleic Acids Res. 50, e60–e60 (2022).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Witten, D. M. Classification and clustering of sequencing data using a Poisson model. Ann. Appl. Stat. 5 (2011).
Liao, Y., Smyth, G. K. & Shi, W. FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Love, M. I., Huber, W. & Anders, S. Moderated Estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Wickham, H. Ggplot2: Elegant Graphics for Data Analysis. (Springer-Verlag, 2016).
Chemello, F. et al. Degenerative and regenerative pathways underlying Duchenne muscular dystrophy revealed by single-nucleus RNA sequencing. Proc. Natl. Acad. Sci. 117, 29691–29701 (2020).
Hänzelmann, S., Castelo, R. & Guinney, J. GSVA: gene set variation analysis for microarray and RNA-Seq data. BMC Bioinform. 14, 7 (2013).
Wu, T. et al. ClusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innov. 2, 100141 (2021).
Korotkevich, G. et al. Fast gene set enrichment analysis. bioRxiv https://doi.org/10.1101/060012 (2021).
Yu, G., Wang, L. G., Yan, G. R. & He, Q. Y. DOSE: an R/Bioconductor package for disease ontology semantic and enrichment analysis. Bioinformatics 31, 608–609 (2015).
Buenrostro, J. D., Giresi, P. G., Zaba, L. C., Chang, H. Y. & Greenleaf, W. J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 10, 1213–1218 (2013).
Buenrostro, J. D., Wu, B., Chang, H. Y. & Greenleaf, W. J. ATAC-seq: A Method for Assaying Chromatin Accessibility Genome‐Wide. Curr Protoc. Mol. Biol 109, https://doi.org/10.1002/0471142727.mb2129s109 (2015).
Young, M. et al. The developmental impacts of natural selection on human pelvic morphology. Sci. Adv. 8 (2022).
Corces, M. R. et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods. 14, 959–962 (2017).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with bowtie 2. Nat. Methods. 9, 357–359 (2012).
Li, H. et al. The sequence Alignment/Map format and samtools. Bioinformatics 25, 2078–2079 (2009).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Lawrence, M. et al. Software for computing and annotating genomic ranges. PLoS Comput. Biol. 9, e1003118 (2013).
Li, Q., Brown, J. B., Huang, H. & Bickel, P. J. Measuring reproducibility of high-throughput experiments. Ann. Appl. Stat. 5 (2011).
Laiker, I. & Frankel, N. Pleiotropic enhancers are ubiquitous regulatory elements in the human genome. Genome Biol. Evol. 14 (2022).
Amemiya, H. M., Kundaje, A. & Boyle, A. P. The ENCODE blacklist: identification of problematic regions of the genome. Sci. Rep. 9, 9354 (2019).
Guo, M. et al. Epigenetic profiling of growth plate chondrocytes sheds insight into regulatory genetic variation influencing height. Elife 6 (2017).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Pollard, K. S., Hubisz, M. J., Rosenbloom, K. R. & Siepel, A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 20, 110–121 (2010).
Karolchik, D. The UCSC genome browser database. Nucleic Acids Res. 31, 51–54 (2003).
Maas, S. A. & Fallon, J. F. Single base pair change in the long-range Sonic Hedgehog limb‐specific enhancer is a genetic basis for preaxial polydactyly. Dev. Dyn. 232, 345–348 (2005).
Prabhakar, S. et al. Human-specific gain of function in a developmental enhancer. Sci. (1979). 321, 1346–1350 (2008).
Wittkopp, P. J. & Kalay, G. Cis -regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat. Rev. Genet. 13, 59–69 (2011).
Richard, D. et al. Evolutionary selection and constraint on human knee chondrocyte regulation impacts osteoarthritis risk. Cell 181, 362–381e28 (2020).
Yu, G., Wang, L. G. & He, Q. Y. ChIPseeker: an R/Bioconductor package for chip peak annotation, comparison and visualization. Bioinformatics 31, 2382–2383 (2015).
McLean, C. Y. et al. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28, 495–501 (2010).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 38, 576–589 (2010).
Bengtsen, M. et al. Comparing the epigenetic landscape in myonuclei purified with a PCM1 antibody from a fast/glycolytic and a slow/oxidative muscle. PLoS Genet. 17, e1009907 (2021).
Vincze, T., Posfai, J. & Roberts, R. J. NEBcutter: a program to cleave DNA with restriction enzymes. Nucleic Acids Res. 31, 3688–3691 (2003).
Maxam, A. M. (ed, W.) A new method for sequencing DNA. Proc. Natl. Acad. Sci. 74 560–564 (1977).
Sun, C. et al. Lineage tracing of nuclei in skeletal myofibers uncovers distinct transcripts and interplay between myonuclear populations. Nat. Commun. 15, 9372 (2024).
Van den Berge, K. et al. Normalization benchmark of ATAC-seq datasets shows the importance of accounting for GC-content effects. Cell. Rep. Methods. 2, 100321 (2022).
Yamaguchi, Y., Kodama, R. & Yamada, S. Morphogenetic progression of thigh and lower leg muscles during human embryonic development. Anat. Rec. 306, 2072–2080 (2023).
Queeno, S. R., Sterner, K. N. & O’Neill, M. C. Meta-analysis data of skeletal muscle slow fiber content across mammalian species. Data Brief. 50, 109520 (2023).
Christ, B. & Brand-Saberi, B. Limb muscle development. Int. J. Dev. Biol. 46, 905–914 (2002).
Ontell, M. P., Sopper, M. M., Lyons, G., Buckingham, M. & Ontell, M. Modulation of contractile protein gene expression in fetal murine crural muscles: emergence of muscle diversity. Dev. Dyn. 198, 203–213 (1993).
Sensiate, L. A. et al. Dact gene expression profiles suggest a role for this gene family in integrating Wnt and TGF-β signaling pathways during chicken limb development. Dev. Dyn. 243, 428–439 (2014).
Hostikka, S. L. & Capecchi, M. R. The mouse Hoxc11 gene: genomic structure and expression pattern. Mech. Dev. 70, 133–145 (1998).
Steingruber, L. et al. ALDH1A1 and ALDH1A3 paralogues of aldehyde dehydrogenase 1 control myogenic differentiation of skeletal muscle satellite cells by retinoic acid-dependent and -independent mechanisms. Cell. Tissue Res. 394, 515–528 (2023).
Lin, X. et al. Hoxa11 and Hoxa13 facilitate slow-twitch muscle formation in C2C12 cells and indirectly affect the lipid deposition of 3T3‐L1 cells. Animal Sci. J. 92, (2021).
de Wilde, J. et al. The embryonic genes Dkk3, Hoxd8, Hoxd9 and Tbx1 identify muscle types in a diet-independent and fiber-type unrelated way. BMC Genom. 11, 176 (2010).
Ono, Y. et al. Scleraxis-lineage cells are required for correct muscle patterning. Development https://doi.org/10.1242/dev.201101 (2023).
Aoto, K. et al. ATP6V0A1 encoding the a1-subunit of the V0 domain of vacuolar H+-ATPases is essential for brain development in humans and mice. Nat. Commun. 12, 2107 (2021).
Ataman, B. et al. Evolution of osteocrin as an activity-regulated factor in the primate brain. Nature 539, 242–247 (2016).
Zito, A. et al. Neuritin 1 promotes neuronal migration. Brain Struct. Funct. 219, 105–118 (2014).
Wang, J. et al. Long Non-coding RNA HOTAIR in Central Nervous System Disorders: New Insights in Pathogenesis, Diagnosis, and Therapeutic Potential. Front. Mol. Neurosci 15, 949095 (2022).
Li, S. M. H. et al. Skin regional specification and higher-order HoxC regulation. Sci. Adv. https://doi.org/10.1126/sciadv.ado2223 (2025).
Gibson-Brown, J. J., Agulnik, S. I., Silver, L. M., Niswander, L. & Papaioannou, V. E. Involvement of T-box genes Tbx2-Tbx5 in vertebrate limb specification and development. Development 125, 2499–2509 (1998).
Sweat, M. E. et al. Tbx5 maintains atrial identity in postnatal cardiomyocytes by regulating an atrial-specific enhancer network. Nat. Cardiovasc. Res. 2, 881–898 (2023).
Cardoso-Moreira, M. et al. Gene expression across mammalian organ development. Nature 571, 505–509 (2019).
La Manno, G. et al. Molecular architecture of the developing mouse brain. Nature 596, 92–96 (2021).
Cai, S. et al. Integrative single-cell RNA-seq and ATAC-seq analysis of myogenic differentiation in pig. BMC Biol. 21, 19 (2023).
Jin, Y. et al. Glutathione S-transferase mu 2 inhibits hepatic steatosis via ASK1 suppression. Commun. Biol. 5, 326 (2022).
O’Reilly, M. E. et al. linc-ADAIN, a human adipose lincRNA, regulates adipogenesis by modulating KLF5 and IL-8 mRNA stability. Cell. Rep. 43, 114240 (2024).
Wang, F., Liang, R., Soibam, B., Yang, J. & Liu, Y. Coregulatory long non-coding RNA and protein-coding genes in serum starved cells. Biochim. Et Biophys. Acta (BBA) - Gene Regul. Mech. 1862, 84–95 (2019).
Scott, T. A., Soemardy, C., Ray, R. M. & Morris, K. V. Targeted zinc-finger repressors to the oncogenic HBZ gene inhibit adult T-cell leukemia (ATL) proliferation. NAR Cancer https://doi.org/10.1093/narcan/zcac046 (2023).
Yamada, M., Warabi, E., Oishi, H., Lira, V. A. & Okutsu, M. Muscle p62 stimulates the expression of antioxidant proteins alleviating cancer cachexia. FASEB J. 37 (2023).
Huraskin, D. et al. Wnt/β-catenin signaling via Axin2 is required for myogenesis and, together with YAP/Taz and Tead1, active in IIa/IIx muscle fibers. Development 143, 3128–3142 (2016).
Hwang, M., Lee, E. J., Chung, M. J., Park, S. & Jeong, K. S. Five transcriptional factors reprogram fibroblast into myogenic lineage cells via paraxial mesoderm stage. Cell. Cycle. 19, 1804–1816 (2020).
Bonczek, O., Balcar, V. J. & Šerý, O. PAX9 gene mutations and tooth agenesis: A review. Clin. Genet. 92, 467–476 (2017).
Mita, Y. et al. R-spondin3 is a myokine that differentiates myoblasts to type I fibres. Sci. Rep. 12, 13020 (2022).
Pan, H. et al. A role for Zic1 and Zic2 in Myf5 regulation and Somite myogenesis. Dev. Biol. 351, 120–127 (2011).
Deng, C. et al. TNFRSF19 inhibits TGFβ signaling through interaction with TGFβ receptor type I to promote tumorigenesis. Cancer Res. 78, 3469–3483 (2018).
Shima, N. et al. Up-regulated expression of two-pore domain K + channels, KCNK1 and KCNK2, is involved in the proliferation and migration of pulmonary arterial smooth muscle cells in pulmonary arterial hypertension. Front Cardiovasc. Med 11, 1343804 (2024).
Jang, D. G., Kwon, K. Y., Song, E. K. & Park, T. J. Integrin β-like 1 protein (ITGBL1) promotes cell migration by preferentially inhibiting integrin-ECM binding at the trailing edge. Genes Genomics. 44, 405–413 (2022).
Lee, W. et al. Role of HIF-1α-Activated IL-22/IL-22R1/Bmi1 signaling modulates the Self-Renewal of cardiac stem cells in acute myocardial ischemia. Stem Cell. Rev. Rep. 20, 2194–2214 (2024).
Javed, A., Abbas, H. B., Ahmed, S., Ahmed, A. & Trali, G. A. In Silico analysis of molecular interactions of FZD10 in Wnt signaling pathway involved in wound healing. Pakistan J. Med. Health Sci. 15, 2841–2844 (2021).
Yu, X., Riley, T. & Levine, A. J. The regulation of the endosomal compartment by p53 the tumor suppressor gene. FEBS J. 276, 2201–2212 (2009).
Bomholt, A. B. et al. Evaluation of commercially available glucagon receptor antibodies and glucagon receptor expression. Commun. Biol. 5, 1278 (2022).
Guo, F. et al. NOTUM promotes thermogenic capacity and protects against diet-induced obesity in male mice. Sci. Rep. 11, 16409 (2021).
Lee, L. A., Karabina, A., Broadwell, L. J. & Leinwand, L. A. The ancient sarcomeric myosins found in specialized muscles. Skelet. Muscle. 9, 7 (2019).
Desjardins, P. R., Burkman, J. M., Shrager, J. B., Allmond, L. A. & Stedman, H. H. Evolutionary implications of three novel members of the human sarcomeric myosin heavy chain gene family. Mol. Biol. Evol. 19, 375–393 (2002).
Zhu, J. et al. Comparative genomics search for losses of Long-Established genes on the human lineage. PLoS Comput. Biol. 3, e247 (2007).
Hoh, J. F. Y. Myosin heavy chains in extraocular muscle fibres: Distribution, regulation and function. Acta Physiologica 231, e13535 (2021).
Klemm, S. L., Shipony, Z. & Greenleaf, W. J. Chromatin accessibility and the regulatory epigenome. Nat. Rev. Genet. 20, 207–220 (2019).
Picardi, E. & Pesole, G. Mitochondrial genomes gleaned from human whole-exome sequencing. Nat. Methods. 9, 523–524 (2012).
Montefiori, L. et al. Reducing mitochondrial reads in ATAC-seq using CRISPR/Cas9. Sci. Rep. 7, 2451 (2017).
Rhodes, C. T. et al. An epigenome atlas of neural progenitors within the embryonic mouse forebrain. Nat. Commun. 13, 4196 (2022).
Abbasova, L. et al. CUT&Tag recovers up to half of ENCODE ChIP-seq histone acetylation peaks. Nat. Commun. 16, 2993 (2025).
Asfour, H. A., Allouh, M. Z. & Said, R. S. Myogenic regulatory factors: the orchestrators of myogenesis after 30 years of discovery. Exp. Biol. Med. 243, 118–128 (2018).
Zhou, P. et al. Identification of novel transcription factors regulated by H3K27 acetylation in myogenic differentiation of porcine skeletal muscle satellite cells. The FASEB Journal 38, e70144 (2024).
Spinelli, S. et al. Estrogen-Related receptor α: A key transcription factor in the regulation of energy metabolism at an organismic level and a target of the ABA/LANCL hormone receptor system. Int. J. Mol. Sci. 25, 4796 (2024).
Lee, H. J. et al. Dysregulation of nuclear receptor COUP-TFII impairs skeletal muscle development. Sci. Rep. 7, 3136 (2017).
Tontonoz, P. et al. The orphan nuclear receptor Nur77 is a determinant of myofiber size and muscle mass in mice. Mol. Cell. Biol. 35, 1125–1138 (2015).
Liu, N. et al. Requirement of MEF2A, C, and D for skeletal muscle regeneration. In Proceedings of the National Academy of Sciences. 111 4109–4114 (2014).
Sanchez, A. M. J., Candau, R. B. & Bernardi, H. FoxO transcription factors: their roles in the maintenance of skeletal muscle homeostasis. Cell. Mol. Life Sci. 71, 1657–1671 (2014).
Sousa-Victor, P., García-Prat, L. & Muñoz-Cánoves, P. Control of satellite cell function in muscle regeneration and its disruption in ageing. Nat. Rev. Mol. Cell. Biol. 23, 204–226 (2022).
Braun, T. & Gautel, M. Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis. Nat. Rev. Mol. Cell. Biol. 12, 349–361 (2011).
Doni Jayavelu, N., Jajodia, A., Mishra, A. & Hawkins, R. D. Candidate silencer elements for the human and mouse genomes. Nat. Commun. 11, 1061 (2020).
Wong, E. S. et al. Deep conservation of the enhancer regulatory code in animals. Science (1979) https://doi.org/10.1126/science.aax8137 (2020).
Orchard, P. et al. Human and rat skeletal muscle single-nuclei multi-omic integrative analyses nominate causal cell types, regulatory elements, and SNPs for complex traits. Genome Res. 31, 2258–2275 (2021).
Nord, A. S. et al. Rapid and pervasive changes in Genome-wide enhancer usage during mammalian development. Cell 155, 1521–1531 (2013).
Thomas, S. et al. Dynamic reprogramming of chromatin accessibility during drosophilaembryo development. Genome Biol. 12, R43 (2011).
Dunwell, T. L. & Holland, P. W. H. Diversity of human and mouse homeobox gene expression in development and adult tissues. BMC Dev. Biol. 16, 40 (2016).
Gluck, C. et al. RNA-seq based transcriptomic map reveals new insights into mouse salivary gland development and maturation. BMC Genom. 17, 923 (2016).
Su, X. et al. Single-cell RNA-Seq analysis reveals dynamic trajectories during mouse liver development. BMC Genom. 18, 946 (2017).
He, P. et al. The changing mouse embryo transcriptome at whole tissue and single-cell resolution. Nature 583, 760–767 (2020).
Petit, F., Sears, K. E. & Ahituv, N. Limb development: a paradigm of gene regulation. Nat. Rev. Genet. 18, 245–258 (2017).
Wong, M. K. et al. Timing of Tissue-specific cell division requires a differential onset of zygotic transcription during metazoan embryogenesis. J. Biol. Chem. 291, 12501–12513 (2016).
Lacombe, J. et al. Genetic and functional modularity of hox activities in the specification of Limb-Innervating motor neurons. PLoS Genet. 9, e1003184 (2013).
Lawrence, J. E. G. et al. HOX gene expression in the developing human spine. Nat. Commun. 15, 10023 (2024).
Capdevila, J. & Belmonte, J. C. I. Patterning mechanisms controlling vertebrate limb development. Annu. Rev. Cell. Dev. Biol. 17, 87–132 (2001).
Hnisz, D. et al. Super-Enhancers in the control of cell identity and disease. Cell 155, 934–947 (2013).
Lei, S. et al. Roles of super enhancers and enhancer RNAs in skeletal muscle development and disease. Cell. Cycle. 22, 495–505 (2023).
Zhang, S. et al. Analyzing super-enhancer Temporal dynamics reveals potential critical enhancers and their gene regulatory networks underlying skeletal muscle development. Genome Res. 34, 2190–2202 (2024).
Casanova, A., Wevers, A., Navarro-Ledesma, S. & Pruimboom, L. Mitochondria: It is all about energy. Front Physiol 14, 1114231 (2023).
Sin, J. et al. Mitophagy is required for mitochondrial biogenesis and myogenic differentiation of C2C12 myoblasts. Autophagy 12, 369–380 (2016).
Smerdu, V. & Cvetko, E. Myosin heavy chain-2b transcripts and isoform are expressed in human laryngeal muscles. Cells Tissues Organs. 198, 75–86 (2013).
Pereira Sant’Ana, J. A., Ennion, S., Sargeant, A. J., Moorman, A. F. & Goldspink, G. Comparison of the molecular, antigenic and ATPase determinants of fast myosin heavy chains in rat and human: a single-fibre study. Pflügers Archive Eur. J. Physiol. 435, 151–163 (1997).
Staron, R. S. Human skeletal muscle fiber types: Delineation, Development, and distribution. Can. J. Appl. Physiol. 22, 307–327 (1997).
Rivero, J. L., Serrano, A. L., Barrey, E., Valette, J. P. & Jouglin, M. Analysis of myosin heavy chains at the protein level in horse skeletal muscle. J. Muscle Res. Cell. Motil. 20, 211–221 (1999).
Harrison, B. C., Allen, D. L. & Leinwand, L. A. IIb or not IIb? Regulation of myosin heavy chain gene expression in mice and men. Skelet. Muscle. 1, 5 (2011).
IJkema-Paassen, J. & Gramsbergen, A. Development of postural muscles and their innervation. Neural Plast. 12, 141–151 (2005).
Pette, D. Historical perspectives: plasticity of mammalian skeletal muscle. J. Appl. Physiol. 90, 1119–1124 (2001).
Wigmore, P. M. & Evans, D. J. R. Molecular and cellular mechanisms involved in the generation of fiber diversity during myogenesis. Int. rev. cytol. 216, 175–232. https://doi.org/10.1016/S0074-7696(02)16006-2 (2002).
Lang, F. et al. Single muscle fiber proteomics reveals distinct protein changes in slow and fast fibers during muscle atrophy. J. Proteome Res. 17, 3333–3347 (2018).
Ahn, J. S. et al. Ectopic overexpression of Porcine Myh1 increased in slow muscle fibers and enhanced endurance exercise in Transgenic mice. Int. J. Mol. Sci. 19, 2959 (2018).
Smerdu, V. Expression of MyHC-15 and ‐2x in human muscle spindles: an immunohistochemical study. J. Anat. 243, 826–841 (2023).
Smerdu, V., Ugwoke, C. K. & Šink, Ž. Co-expression of MyHC-15 with other known isoforms in rat muscle spindles. European J. Histochemistry 69, 4192 (2025).
Rossi, A. C., Mammucari, C., Argentini, C., Reggiani, C. & Schiaffino, S. Two novel/ancient myosins in mammalian skeletal muscles: MYH14/7b and MYH15 are expressed in extraocular muscles and muscle spindles. J. Physiol. 588, 353–364 (2010).
Yang, J. H. & Hansen, A. S. Enhancer selectivity in space and time: from enhancer–promoter interactions to promoter activation. Nat. Rev. Mol. Cell. Biol. 25, 574–591 (2024).
Lawrence, M., Daujat, S. & Schneider, R. Lateral thinking: how histone modifications regulate gene expression. Trends Genet. 32, 42–56 (2016).
Oe, M., Ojima, K. & Muroya, S. Difference in potential DNA methylation impact on gene expression between fast- and slow-type myofibers. Physiol. Genomics. 53, 69–83 (2021).
Wang, B., Starr, A. L. & Fraser, H. B. Cell-type-specific cis-regulatory divergence in gene expression and chromatin accessibility revealed by human-chimpanzee hybrid cells. Elife https://doi.org/10.7554/eLife.89594 (2024).
Reilly, S. K. & Noonan, J. P. Evolution of gene regulation in humans. Annu. Rev. Genomics Hum. Genet. 17, 45–67 (2016).
Tang, F. et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods. 6, 377–382 (2009).
Buenrostro, J. D. et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523, 486–490 (2015).
Giordani, L. et al. High-Dimensional Single-Cell cartography reveals novel skeletal Muscle-Resident cell populations. Mol. Cell. 74, 609–621e6 (2019).
Rubenstein, A. B. et al. Single-cell transcriptional profiles in human skeletal muscle. Sci. Rep. 10, 229 (2020).
Hollingsworth, E. W. et al. Rapid and quantitative functional interrogation of human enhancer variant activity in live mice. Nat. Commun. 16, 409 (2025).
Chang, T. Y. & Waxman, D. J. HDI-STARR-seq: Condition-specific enhancer discovery in mouse liver in vivo. BMC Genom. 25, 1240 (2024).
Osterwalder, M. et al. Characterization of Mammalian In Vivo Enhancers Using Mouse Transgenesis and CRISPR Genome Editing. Craniofacial Dev. Methods Protocols https://doi.org/10.1007/978-1-0716-1847-9_11 (2022).
Lambert, J. T. et al. Parallel functional testing identifies enhancers active in early postnatal mouse brain. Elife 10, e69479 (2021).
Pennacchio, L. A. et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature 444, 499–502 (2006).
Acknowledgements
This study was supported by the University of Oregon, National Science Foundation (SRQ BCS-1945809 and ASO DGE-1745303) and the Leakey Foundation. Thanks to Emmanuelle Boucicaut, Joseph Braud, Jack Chambers, Emma Freedman and Elijah Reed for their help in the cell culture lab and to Natalie Dunn, Carrie McCurdy, Kristin Kohler, Daniel Richard, Avika Gomez-Sharma, Allissa Van Steenis and Mariel Young for their expertise and technical support. This work benefited from access to the University of Oregon high performance computing cluster, Talapas. Some figures were created using BioRender.
Author information
Authors and Affiliations
Contributions
SRQ, TDC, and KNS conceptualized the study; KNS and TDC supervised the project; ASO and SRQ performed the experiments; SRQ wrote the manuscript, analyzed the data, and prepared the figures; MCO and DMC substantially revised the manuscript for intellectual content, and all authors edited the manuscript. All authors approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Queeno, S.R., Okamoto, A.S., Callahan, D.M. et al. Profiling the epigenomic landscape of late embryonic and adult mouse hind limb muscles. Sci Rep (2026). https://doi.org/10.1038/s41598-025-32705-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-32705-4