Abstract
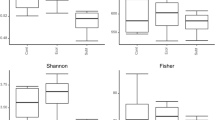
Freshwater ecosystems are vital for biodiversity and livelihoods in Bangladesh, where interest in fish gut microbiota is growing to aid aquaculture sustainability through microbial interventions. Therefore, this research investigated the bacteriomes of the gut of Rohu from the Halda River and Kaptai Lake, using Oxford Nanopore long-read 16S rRNA sequencing. The evaluation of diversity demonstrated notable variations in both alpha and beta diversity indices (p < 0.05). The fish in the Halda River had a varied bacteriome, mostly composed of Pirellulaceae_uncultured (9.26%), with environmentally tolerant taxa such as Exiguobacterium (5.48%). In contrast, the Kaptai Lake fish have a bacteriome that is abundant in probiotics, including Lactiplantibacillus (48.84%) and Pediococcus (8.82%). Water samples exhibited unique microbiological signatures: Halda River water was mostly characterized by Exiguobacterium (41.93%), while Kaptai Lake water was primarily composed of Acinetobacter (71.24%). Furthermore, functional analysis indicated that fish from the Halda River comprised metabolically diverse communities involved in nitrogen cycling, whereas the Kaptai Lake fish demonstrated a strong capacity for ammonia oxidation and pollutant breakdown. The research offers significant insights into the relationship between the host, microbiome, and environment, with implications for enhancing fish health and promoting sustainable aquaculture practices.
Similar content being viewed by others
Data availability
Sequence data that support the findings of this study have been deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) with the primary accession code [PRJNA1270017](https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1270017) .
References
DOF. Yearbook of Fisheries Statistics of Bangladesh, 2022–23. 40, 138 (Fisheries Resource Survey System, Ministry of Fisheries and Livestock, 2023).
Ferdous, M., Karim, M., Hossain, M., Rahman, M. & Iqbal, M. Fin fish assemblage and biodiversity status of carps on Halda River. Bangladesh. Ann. Vet. Anim. Sci. 2, 151–161 (2015).
Arshad-Ul-Alam, M. & Azadi, M. Fisheries exploitation of the Halda River, Bangladesh. J. Fish. 4, 361–370 (2016).
Nath, R. K. et al. Assessment of heavy metal concentration in the water of major carp breeding River Halda Bangladesh. Sustain. Chem. One World. 3, 100018 (2024). https://doi.org/10.1016/j.scowo.2024.100018.
Patra, R. & Azadi, M. Hydrological conditions influencing the spawning of major carps in the Halda River, Chittagong, Bangladesh. Bangladesh J. Zool. 13, 63–72 (1985).
Shampa, M. T. A., Shimu, N. J., Chowdhury, K. M. A., Islam, M. M. & Ahmed, M. K. A comprehensive review on sustainable coastal zone management in Bangladesh: present status and the way forward. Heliyon 9, 18190. https://doi.org/10.1016/j.heliyon.2023.e18190 (2023).
Boyd, C. E. et al. Achieving sustainable aquaculture: historical and current perspectives and future needs and challenges. J. World Aquac. Soc. 51, 578–633 (2020).
Lima, R. et al. An overview on hydro-biology and management of Kaptai Lake fisheries. Bangladesh. Int. J. Aquac. Fish. Sci. 9, 29–39 (2023).
Karmakar, S., Haque, S. S., Hossain, M. & Shafiq, M. Water quality of Kaptai reservoir in Chittagong Hill tracts of Bangladesh. J. For. Res. 22, 87–92 (2011).
Ahmed, K., Rahman, S. & Ahammed, S. Managing fisheries resources in Kaptai Reservoir, Bangladesh. Outlook Agric. 35, 281–289 (2006).
Naher, S. & Galib, S. Fishes of Kaptai Lake: management and conservation perspectives. J. Life Earth Sci. 15, 53–58 (2020).
Arafeen, M. et al. Present status of fish diversity of the Kaptai Lake. J. Bangladesh Agric. Univ. 22, 506–516 (2024).
Bakar, M. & Bhuyan, M. Assessment of water quality in Halda River (the major carp breeding ground) of Bangladesh. J. Pollut. 3, 429–444 (2017).
Talwar, C., Nagar, S., Lal, R. & Negi, R. K. Fish gut microbiome: Current approaches and future perspectives. Indian J. Microbiol. 58, 397–414 (2018).
Kanika, N. H. et al. Fish gut microbiome and its application in aquaculture and biological conservation. Front. Microbiol. 15, 1521048. https://doi.org/10.3389/fmicb.2024.1521048 (2025).
See, M. S. et al. Aquatic microbiomes under stress: The role of gut microbiota in detoxification and adaptation to environmental exposures. J. Hazard. Mater. Adv. 17, 100612. https://doi.org/10.1016/j.hazadv.2025.100612 (2025).
Stephens, W. Z. et al. The composition of the zebrafish intestinal microbial community varies across development. ISME J. 10, 644–654 (2016).
Li, X. et al. Gut microbiota contributes to the growth of fast-growing transgenic common carp (Cyprinus carpio L.). PLoS ONE 8, 64577. https://doi.org/10.1371/journal.pone.0064577 (2013).
Giatsis, C. et al. Probiotic legacy effects on gut microbial assembly in tilapia larvae. Sci. Rep. 6, 33965. https://doi.org/10.1038/srep33965 (2016).
Nayak, S. K. Role of gastrointestinal microbiota in fish. Aquac. Res. 41, 1553–1573 (2010).
Jyoti, & Dey, P. Mechanisms and implications of the gut microbial modulation of intestinal metabolic processes. Npj Metab. Health Dis. 3, 24 (2025).
Pham, V. T., Dold, S., Rehman, A., Bird, J. K. & Steinert, R. E. Vitamins, the gut microbiome and gastrointestinal health in humans. Nutr. Res. 95, 35–53 (2021).
Llewellyn, M. S., Boutin, S., Hoseinifar, S. H. & Derome, N. Teleost microbiomes: The state of the art in their characterization, manipulation and importance in aquaculture and fisheries. Front. Microbiol. 5, 207 (2014).
Lee, P.-T., Yamamoto, F. Y., Low, C.-F., Loh, J.-Y. & Chong, C.-M. Gut immune system and the implications of oral-administered immunoprophylaxis in finfish aquaculture. Front. Immunol. 12, 773193. https://doi.org/10.3389/fimmu.2021.773193 (2021).
Linda, S. S. et al. Synbiotic supplementation boosts growth, gut health, and immunity in Asian fossil catfish (Heteropneustes fossilis). Aquac. Res. 2025, 4542077. https://doi.org/10.1155/are/4542077 (2025).
Ringø, E. et al. Effect of dietary components on the gut microbiota of aquatic animals: A never-ending story?. Anim. Nutr. 22, 219–282 (2016).
El-Saadony, M. T. et al. The functionality of probiotics in aquaculture: an overview. Fish Shellfish Immunol. 117, 36–52 (2021).
Sullam, K. E. et al. Environmental and ecological factors that shape the gut bacterial communities of fish: A meta-analysis. Mol. Ecol. 21, 3363–3378 (2012).
Egerton, S., Culloty, S., Whooley, J., Stanton, C. & Ross, R. P. The gut microbiota of marine fish. Front. Microbiol. 9, 873 (2018).
Maji, U. et al. Exploring the gut microbiota composition of Indian major carp, rohu (Labeo rohita), under diverse culture conditions. Genomics 114, 110354. https://doi.org/10.1016/j.ygeno.2022.110354 (2022).
Cardona, E. et al. Bacterial community characterization of water and intestine of the shrimp Litopenaeus stylirostris in a biofloc system. BMC Microbiol. 16, 157 (2016).
Caporaso, J. G. et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624 (2012).
Percie du Sert, N. et al. The arrive guidelines 20: Updated guidelines for reporting animal research. PLoS Biol. 18, 3000410. https://doi.org/10.1371/journal.pbio.3000410 (2020).
Klindworth, A. et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41, e1. https://doi.org/10.1093/nar/gks808 (2013).
Bonenfant, Q., Noé, L. & Touzet, H. Porechop_ABI: Discovering unknown adapters in Oxford Nanopore technology sequencing reads for downstream trimming. Bioinform. Adv. 3, 085. https://doi.org/10.1093/bioadv/vbac085 (2022).
De Coster, W., D’Hert, S., Schultz, D. T., Cruts, M. & Van Broeckhoven, C. NanoPack: Visualizing and processing long-read sequencing data. Bioinformatics 34, 2666–2669 (2018).
Tyler, A. D. et al. Evaluation of Oxford Nanopore’s MinION sequencing device for microbial whole genome sequencing applications. Sci. Rep. 8, 10931. https://doi.org/10.1038/s41598-018-29334-5 (2018).
Quast, C. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, 590–596 (2013).
Wood, D. E., Lu, J. & Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 20, 257. https://doi.org/10.1186/s13059-019-1891-0 (2019).
McMurdie, P. J. & Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8, 61217. https://doi.org/10.1371/journal.pone.0061217 (2013).
Oksanen, J. et al. Vegan: community ecology package. R Package Version 2.2–1, 1–2 https://cran.r-project.org/web/packages/vegan/vegan.pdf (2015).
Arndt, D. et al. METAGENassist: A comprehensive web server for comparative metagenomics. Nucleic Acids Res. 40, 88–95 (2012).
Ringø, E. et al. Lactic acid bacteria in finfish—An update. Front. Microbiol. 9, 1818. https://doi.org/10.3389/fmicb.2018.01818 (2018).
Salma, U. et al. Occurrence, risks, and mitigation of antibiotic pollution in Bangladeshi aquaculture systems. Environ. Chem. Ecotoxicol. 7, 351–363 (2025).
Okeke, E. S. et al. Antibiotic resistance in aquaculture and aquatic organisms: a review of current nanotechnology applications for sustainable management. Environ. Sci. Pollut. Res. Int. 29, 69241–69274 (2022).
Popoola, B. M., Adeyemi, O. A. & Samson, O. J. Antibiotic-resistant bacteria in tropical freshwater ecosystems: A review of occurrence, distribution and environmental implications. Microbe https://doi.org/10.1016/j.microb.2025.100457 (2025).
Buckland, S. T., Magurran, A. E., Green, R. E. & Fewster, R. M. Monitoring change in biodiversity through composite indices. Phil. Trans. R. Soc. 360, 243–254 (2005).
Yasmin, Z. et al. Comparison of fecal bacteriome of diarrhoeic and non-diarrhoeic calves revealed diversified community structures. Microbe 6, 100251. https://doi.org/10.1016/j.microb.2025.100251 (2025).
Newton, R. J., Jones, S. E., Eiler, A., McMahon, K. D. & Bertilsson, S. A guide to the natural history of freshwater lake bacteria. Microbiol. Mol. Biol. Rev. 75, 14–49 (2011).
Liu, T.-T. & Yang, H. Comparative analysis of the total and active bacterial communities in the surface sediment of Lake Taihu. FEMS Microbiol. Ecol. 96, 059. https://doi.org/10.1093/femsec/fiaa059 (2020).
Vitorino, I. R., Gambardella, N., Semedo, M., Magalhães, C. & Lage, O. M. Diversity and vertical distribution of Planctomycetota in the water column of the remote North Pacific. Environ. Microbiol. Rep. 17, 70063. https://doi.org/10.1111/1758-2229.70063 (2025).
Kaboré, O. D., Godreuil, S. & Drancourt, M. Planctomycetes as host-associated bacteria: a perspective that holds promise for their future isolations, by mimicking their native environmental niches in clinical microbiology laboratories. Front. Cell. Infect. Microbiol. 10, 519301. https://doi.org/10.3389/fcimb.2020.519301 (2020).
Katiku, M. M., Matofari, J. W. & Nduko, J. M. Preliminary evaluation of probiotic properties and safety profile of Lactiplantibacillus plantarum isolated from spontaneously fermented milk, Amabere amaruranu. Heliyon 8, 10342. https://doi.org/10.1016/j.heliyon.2022.e10342 (2022).
Kumari, V. B. et al. Characterization of Lactobacillus spp. as probiotic and antidiabetic potential isolated from Boza, traditional fermented beverage in Turkey. Adv. Biol. 2024, 2148676. https://doi.org/10.1155/2024/2148676 (2024).
Xie, G. et al. Functional genomic characterization reveals the probiotic tendency and safety assessment of Exiguobacterium acetylicum G1–33 isolated from the gut of the hybrid grouper (Epinephelus fuscoguttatus♀ × E. lanceolatus♂). Aquac. Rep. 36, 102127; https://doi.org/10.1016/j.aqrep.2024.102127 (2024).
Bai, J. et al. Different lactic acid bacteria and their combinations regulated the fermentation process of ensiled alfalfa: ensiling characteristics, dynamics of bacterial community and their functional shifts. Microb. Biotechnol. 14, 1171–1182 (2021).
Xiong, J. B., Nie, L. & Chen, J. Current understanding on the roles of gut microbiota in fish disease and immunity. Zool. Res. 40, 70–76 (2019).
Luan, Y. et al. The fish microbiota: Research progress and potential applications. Engineering 29, 137–146 (2023).
Mohammed, E. A. H., Ahmed, A. E. M., Kovács, B. & Pál, K. The significance of probiotics in aquaculture: A review of research trend and latest scientific findings. Antibiotics 14, 302. https://doi.org/10.3390/antibiotics14030242 (2025).
Zhang, Y., Shi, P. & Ma, J. Exiguobacterium spp. and their applications in environmental remediation. Chin. J. Appl. Environ. Biol. 19, 898–904 (2013).
Delegan, Y. et al. Characterization and genomic analysis of Exiguobacterium alkaliphilum B-3531D, an efficient crude oil degrading strain. Biotechnol. Rep. 32, 00678. https://doi.org/10.1016/j.btre.2021.e00678 (2021).
Howard, A., O’Donoghue, M., Feeney, A. & Sleator, R. D. Acinetobacter baumannii: An emerging opportunistic pathogen. Virulence 3, 243–250 (2012).
Hossain, M. et al. Mixed dye degradation by Bacillus pseudomycoides and Acinetobacter haemolyticus isolated from industrial effluents: a combined affirmation with wetlab and in silico studies. Arab. J. Chem. 15, 104078. https://doi.org/10.1016/j.arabjc.2022.104078 (2022).
Jung, J. & Park, W. Acinetobacter species as model microorganisms in environmental microbiology: Current state and perspectives. Appl. Microbiol. Biotechnol. 99, 2533–2548 (2015).
Benoit, T. et al. Acinetobacter calcoaceticus-baumannii complex prevalence, spatial-temporal distribution, and contamination sources in Canadian aquatic environments. Microbiol. Spectr. 12, 0150924. https://doi.org/10.1128/spectrum.01509-24 (2024).
Segura, A., Hernández Sánchez, V., Marqués, S. & Molina, L. Insights in the regulation of the degradation of PAHs in Novosphingobium sp HR1a and utilization of this regulatory system as a tool for the detection of PAHs. Sci. Total Environ. 590–591, 381–393 (2017).
Segura, A., Udaondo, Z. & Molina, L. PahT regulates carbon fluxes in Novosphingobium sp HR1a and influences its survival in soil and rhizospheres. Environ. Microbiol. 23, 2969–2991 (2021).
Dong, Y. et al. Distinct gut microbial communities and functional predictions in divergent ophiuroid species: host differentiation, ecological niches, and adaptation to cold-water habitats. Microbiol. Spectr. 11, 0207323. https://doi.org/10.1128/spectrum.02073-23 (2023).
Magnuson, E., Altshuler, I., Freyria, N. J., Leveille, R. J. & Whyte, L. G. Sulfur-cycling chemolithoautotrophic microbial community dominates a cold, anoxic, hypersaline Arctic spring. Microbiome 11, 203. https://doi.org/10.1186/s40168-023-01628-5 (2023).
Hou, Y., Jia, R., Ji, P., Li, B. & Zhu, J. Organic matter degradation and bacterial communities in surface sediment influenced by Procambarus clarkia. Front. Microbiol. 13, 985555. https://doi.org/10.3389/fmicb.2022.985555 (2022).
Hayatsu, M., Tago, K. & Saito, M. Various players in the nitrogen cycle: Diversity and functions of the microorganisms involved in nitrification and denitrification. J. Plant Nutr. Soil Sci. 54, 33–45 (2008).
Omori, T., Iwata, K., Yu, S. S. & Azlan, N. N. A. Ammonia Accumulation of Novel Nitrogen-Fixing Bacteria in Biotechnology—Molecular Studies and Novel Applications for Improved Quality of Human Life (ed Sammour, R. H.) (IntechOpen, 2012).
Wang, X. et al. Adaptation and assembly of microbial communities under saline-alkaline stress in paddy ecosystems: implications for nitrogen and carbon cycling. Environ. Technol. Innov. 39, 104329. https://doi.org/10.1016/j.eti.2025.104329 (2025).
Zhang, K. et al. Fish growth enhances microbial sulfur cycling in aquaculture pond sediments. Microb. Biotechnol. 13, 1597–1610 (2020).
Acknowledgements
We are thankful to the Research and Extension, and Department of Genomics and Bioinformatics, Chattogram Veterinary and Animal Sciences University.
Funding
This research is funded by the Research and Extension (Grant ID: 830-75) Division of Chattogram Veterinary and Animal Sciences University, Chattogram.
Author information
Authors and Affiliations
Contributions
M. S. U., K. C. and M. H. U. M. Conceptualized and designed the study outline, performed all the analyses, prepared the figures, wrote and revised the manuscript; M. R. N., S. C., A. S. and I. H. performed the analyses, wrote the manuscript; M. S. U., M. H. U. M. and ASM. L. A. Supervised the study and revised the manuscript; All authors have agreed with the manuscript and provided their consent for publication.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
The Ethics Approval Committee (EAC) of Chattogram Veterinary and Animal Sciences University (CVASU) reviewed and authorized the study protocol (Approval no. CVASU/Dir (R&E) EC/2025/881/15).
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Uddin, M.S., Chamonara, K., Nayem, M.R. et al. Comparative gut microbiome analysis of Rohu fish from Halda River and Kaptai Lake using 16S rRNA sequencing. Sci Rep (2026). https://doi.org/10.1038/s41598-025-33754-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-33754-5