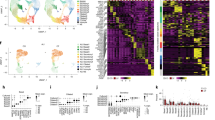
Abstract
Cystic fibrosis (CF) lung disease is characterized by the presence of marked, neutrophil-dominant inflammation that contributes to tissue injury and the development of irreversible structural lung disease. Here, we describe a dysregulated, neutrophil-dominant inflammation and an accompanying pro-inflammatory airway epithelium in the pediatric CF lung through the application of single-cell RNA sequencing (scRNA-seq) to minimally invasive respiratory specimens collected during flexible bronchoscopy. These findings were present in both an infant and an adolescent with CF, the latter on cystic fibrosis transmembrane conductance regulator (CFTR) modulator therapy, suggesting a common pathological program that starts early in life and may be challenging to reverse once structural lung disease is established. Intercellular communication network analysis further revealed potential mechanisms whereby airway epithelial cells modulate the ongoing, destructive airway inflammation present in the CF lung. Importantly, the scRNA-seq workflow leveraged in this study provides a unique opportunity to investigate and monitor disease-related changes in the composition, function, and interaction of the immune and airway epithelial cell populations in CF and other respiratory diseases across the life course.
Similar content being viewed by others
Data availability
Raw FASTQ and processed matrix data that support the findings of this study have been deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus at the National Institutes of Health with the accession number GSE271984. Additional data and code required to analyze the results and generate the figures included in this manuscript are available from the corresponding authors upon reasonable request.
References
Bedrossian, C. W. et al. The lung in cystic fibrosis. A quantitative study including prevalence of pathologic findings among different age groups. Hum. Pathol. 7, 195–204 (1976).
Esterly, J. R. & Oppenheimer, E. H. Observations in cystic fibrosis of the pancreas. 3. Pulmonary lesions. Johns. Hopkins Med. J. 122, 94–101 (1968).
Khan, T. Z. et al. Early pulmonary inflammation in infants with cystic fibrosis. Am. J. Respir Crit. Care Med. 151, 1075–1082 (1995).
Armstrong, D. S. et al. Lower airway inflammation in infants and young children with cystic fibrosis. Am. J. Respir Crit. Care Med. 156, 1197–1204 (1997).
Sly, P. D. et al. Lung disease at diagnosis in infants with cystic fibrosis detected by newborn screening. Am. J. Respir Crit. Care Med. 180, 146–152 (2009).
Martinez, T. M. et al. High-resolution computed tomography imaging of airway disease in infants with cystic fibrosis. Am. J. Respir Crit. Care Med. 172, 1133–1138 (2005).
Stick, S. M. et al. Bronchiectasis in infants and preschool children diagnosed with cystic fibrosis after newborn screening. J. Pediatr. 155, 623–628 (2009).
Sly, P. D. et al. Risk factors for bronchiectasis in children with cystic fibrosis. N Engl. J. Med. 368, 1963–1970 (2013).
Ranganathan, S. C. et al. Airway function in infants newly diagnosed with cystic fibrosis. Lancet 358, 1964–1965 (2001).
Ranganathan, S. C. et al. The evolution of airway function in early childhood following clinical diagnosis of cystic fibrosis. Am. J. Respir Crit. Care Med. 169, 928–933 (2004).
Linnane, B. M. et al. Lung function in infants with cystic fibrosis diagnosed by newborn screening. Am. J. Respir Crit. Care Med. 178, 1238–1244 (2008).
Belessis, Y. et al. Early cystic fibrosis lung disease detected by Bronchoalveolar lavage and lung clearance index. Am. J. Respir Crit. Care Med. 185, 862–873 (2012).
Schupp, J. C. et al. Single-cell transcriptional archetypes of airway inflammation in cystic fibrosis. Am. J. Respir Crit. Care Med. 202, 1419–1429 (2020).
Li, X. et al. ScRNA-seq expression of IFI27 and APOC2 identifies four alveolar macrophage superclusters in healthy BALF. Life Sci. Alliance. 5, e202201458 (2022).
Maksimovic, J. et al. Single-cell atlas of bronchoalveolar lavage from preschool cystic fibrosis reveals new cell phenotypes. Preprint at bioRxiv 2022.2006.2017.496207 (2022).
Berg, M. et al. Evidence for altered immune-structural cell crosstalk in cystic fibrosis revealed by single cell transcriptomics. J Cyst. Fibros (2025).
Carraro, G. et al. Transcriptional analysis of cystic fibrosis airways at single-cell resolution reveals altered epithelial cell States and composition. Nat. Med. 27, 806–814 (2021).
Januska, M. N. et al. Moving toward personalized cystic fibrosis care: a workflow for single-cell RNA sequencing. Am. J. Respir Crit. Care Med. 205, A5179 (2022).
Wauters, E. et al. Discriminating mild from critical COVID-19 by innate and adaptive immune single-cell profiling of Bronchoalveolar lavages. Cell. Res. 31, 272–290 (2021).
Sikkema, L. et al. An integrated cell atlas of the lung in health and disease. Nat. Med. 29, 1563–1577 (2023).
Deprez, M. et al. A single-cell atlas of the human healthy airways. Am. J. Respir Crit. Care Med. 202, 1636–1645 (2020).
Liu, Y. Identification and comprehensive analysis of super-enhancer related genes involved in epithelial-to-mesenchymal transition in lung adenocarcinoma. PLoS One. 18, e0291088 (2023).
Thursfield, R. et al. S82 Airway inflammation is present by 4 months in CF infants diagnosed on newborn screening. Thorax 67, A40–A41 (2012).
Rozhkova, E. et al. Exogenous mammalian extracellular HSP70 reduces endotoxin manifestations at the cellular and organism levels. Ann. N Y Acad. Sci. 1197, 94–107 (2010).
Vinokurov, M. et al. Recombinant human Hsp70 protects against Lipoteichoic acid-induced inflammation manifestations at the cellular and organismal levels. Cell. Stress Chaperones. 17, 89–101 (2012).
Yao, Y. W. et al. Lipopolysaccharide pretreatment protects against ischemia/reperfusion injury via increase of HSP70 and Inhibition of NF-κB. Cell. Stress Chaperones. 16, 287–296 (2011).
Afrazi, A. et al. Intracellular heat shock protein-70 negatively regulates TLR4 signaling in the newborn intestinal epithelium. J. Immunol. 188, 4543–4557 (2012).
Jog, N. R. et al. Heat shock protein 27 regulates neutrophil chemotaxis and exocytosis through two independent mechanisms. J. Immunol. 178, 2421–2428 (2007).
Hashiguchi, N. et al. Enhanced expression of heat shock proteins in activated polymorphonuclear leukocytes in patients with sepsis. J. Trauma. 51, 1104–1109 (2001).
Gupta, S. et al. Heat-shock protein-90 prolongs septic neutrophil survival by protecting c-Src kinase and caspase-8 from proteasomal degradation. J. Leukoc. Biol. 103, 933–944 (2018).
Zheng, L. et al. Pathogen-induced apoptotic neutrophils express heat shock proteins and elicit activation of human macrophages. J. Immunol. 173, 6319–6326 (2004).
Ariga, M. et al. Nonredundant function of phosphodiesterases 4D and 4B in neutrophil recruitment to the site of inflammation. J. Immunol. 173, 7531–7538 (2004).
Cao, M. et al. Mechanisms of impaired neutrophil migration by MicroRNAs in myelodysplastic syndromes. J. Immunol. 198, 1887–1899 (2017).
Xu, X. et al. GPCR-mediated PLCβγ/PKCβ/PKD signaling pathway regulates the Cofilin phosphatase slingshot 2 in neutrophil chemotaxis. Mol. Biol. Cell. 26, 874–886 (2015).
Tang, W. et al. A PLCβ/PI3Kγ-GSK3 signaling pathway regulates Cofilin phosphatase slingshot2 and neutrophil polarization and chemotaxis. Dev. Cell. 21, 1038–1050 (2011).
Liu, X. et al. Single-cell RNA transcriptomic analysis identifies Creb5 and CD11b-DCs as regulator of asthma exacerbations. Mucosal Immunol. 15, 1363–1374 (2022).
Chang, M. et al. Peli1 facilitates TRIF-dependent Toll-like receptor signaling and Proinflammatory cytokine production. Nat. Immunol. 10, 1089–1095 (2009).
Alexis, N. E. et al. Attenuation of host defense function of lung phagocytes in young cystic fibrosis patients. J. Cyst. Fibros. 5, 17–25 (2006).
Zilionis, R. et al. Single-cell transcriptomics of human and mouse lung cancers reveals conserved myeloid populations across individuals and species. Immunity 50, 1317–1334 (2019).
Ratjen, F. et al. Differential cytology of Bronchoalveolar lavage fluid in normal children. Eur. Respir J. 7, 1865–1870 (1994).
Midulla, F. et al. Bronchoalveolar lavage studies in children without parenchymal lung disease: cellular constituents and protein levels. Pediatr. Pulmonol. 20, 112–118 (1995).
Hieshima, K. et al. A novel human CC chemokine PARC that is most homologous to macrophage-inflammatory protein-1 alpha/LD78 alpha and chemotactic for T lymphocytes, but not for monocytes. J. Immunol. 159, 1140–1149 (1997).
Zhan, T. et al. Nicotinamide phosphoribose transferase facilitates macrophage-mediated pulmonary fibrosis through the Sirt1-Smad7 pathway in mice. Eur. J. Pharmacol. 967, 176355 (2024).
Hong, S. M. et al. NAMPT mitigates colitis severity by supporting redox-sensitive activation of phagocytosis in inflammatory macrophages. Redox Biol. 50, 102237 (2022).
Liu, Y. et al. Long non-coding RNA NEAT1 participates in ventilator-induced lung injury by regulating miR-20b expression. Mol. Med. Rep. 25, 66 (2022).
Chen, K. & Kolls, J. K. T cell-mediated host immune defenses in the lung. Annu. Rev. Immunol. 31, 605–633 (2013).
Ingersoll, S. A. et al. Mature cystic fibrosis airway neutrophils suppress T cell function: evidence for a role of arginase 1 but not programmed death-ligand 1. J. Immunol. 194, 5520–5528 (2015).
Jacobsen, L. C. et al. Arginase 1 is expressed in myelocytes/metamyelocytes and localized in gelatinase granules of human neutrophils. Blood 109, 3084–3087 (2007).
Rotondo, R. et al. Exocytosis of azurophil and arginase 1-containing granules by activated polymorphonuclear neutrophils is required to inhibit T lymphocyte proliferation. J. Leukoc. Biol. 89, 721–727 (2011).
Munder, M. et al. Suppression of T-cell functions by human granulocyte arginase. Blood 108, 1627–1634 (2006).
Zea, A. H. et al. L-Arginine modulates CD3zeta expression and T cell function in activated human T lymphocytes. Cell. Immunol. 232, 21–31 (2004).
Regamey, N. et al. Distinct patterns of inflammation in the airway lumen and bronchial mucosa of children with cystic fibrosis. Thorax 67, 164–170 (2012).
Hubeau, C. et al. Quantitative analysis of inflammatory cells infiltrating the cystic fibrosis airway mucosa. Clin. Exp. Immunol. 124, 69–76 (2001).
Hynes, J. et al. Innate immunity in cystic fibrosis: varied effects of CFTR modulator therapy on cell-to-cell communication. Int. J. Mol. Sci. 26, 2636 (2025).
Bingle, L. et al. BPIFB1 (LPLUNC1) is upregulated in cystic fibrosis lung disease. Histochem. Cell. Biol. 138, 749–758 (2012).
Cowland, J. B. & Borregaard, N. Molecular characterization and pattern of tissue expression of the gene for neutrophil gelatinase-associated Lipocalin from humans. Genomics 45, 17–23 (1997).
Cowland, J. B. et al. Neutrophil gelatinase-associated Lipocalin is up-regulated in human epithelial cells by IL-1 beta, but not by TNF-alpha. J. Immunol. 171, 6630–6639 (2003).
Zughaier, S. M. et al. Peripheral monocytes derived from patients with cystic fibrosis and healthy donors secrete NGAL in response to Pseudomonas aeruginosa infection. J. Investig Med. 61, 1018–1025 (2013).
Oglesby, I. K. et al. miR-17 overexpression in cystic fibrosis airway epithelial cells decreases interleukin-8 production. Eur. Respir J. 46, 1350–1360 (2015).
Maiuri, L. et al. Tissue transglutaminase activation modulates inflammation in cystic fibrosis via PPARgamma down-regulation. J. Immunol. 180, 7697–7705 (2008).
Gao, J. et al. Gene interfered-ferroptosis therapy for cancers. Nat. Commun. 12, 5311 (2021).
Luo, T. et al. Ferroptosis assassinates tumor. J. Nanobiotechnol. 20, 467 (2022).
Tang, D. et al. The molecular machinery of regulated cell death. Cell. Res. 29, 347–364 (2019).
Li, Y. et al. Multifaceted roles of ferroptosis in lung diseases. Front. Mol. Biosci. 9, 919187 (2022).
Maniam, P. et al. Increased susceptibility of cystic fibrosis airway epithelial cells to ferroptosis. Biol. Res. 54, 38 (2021).
Sugimoto, M. A. et al. Annexin A1 and the resolution of inflammation: modulation of neutrophil recruitment, apoptosis, and clearance. J. Immunol. Res. 2016, 8239258 (2016).
Bensalem, N. et al. Down-regulation of the anti-inflammatory protein Annexin A1 in cystic fibrosis knock-out mice and patients. Mol. Cell. Proteom. 4, 1591–1601 (2005).
D’Acquisto, F. et al. Pro-inflammatory and pathogenic properties of Annexin-A1: the whole is greater than the sum of its parts. Biochem. Pharmacol. 85, 1213–1218 (2013).
Abouelasrar Salama, S. et al. Serum amyloid A1 (SAA1) revisited: restricted leukocyte-activating properties of homogeneous SAA1. Front. Immunol. 11, 843 (2020).
Abouelasrar Salama, S. et al. Acute-serum amyloid A and A-SAA-derived peptides as formyl peptide receptor (FPR) 2 ligands. Front. Endocrinol. 14, 1119227 (2023).
Dustin, M. L. Complement receptors in myeloid cell adhesion and phagocytosis. Microbiol Spectr 4, 56 (2016).
Stahl, P. L. et al. Visualization and analysis of gene expression in tissue sections by Spatial transcriptomics. Science 353, 78–82 (2016).
Zhu, J. et al. Delineating the dynamic evolution from preneoplasia to invasive lung adenocarcinoma by integrating single-cell RNA sequencing and Spatial transcriptomics. Exp. Mol. Med. 54, 2060–2076 (2022).
Wang, Y. et al. Spatial transcriptomics delineates molecular features and cellular plasticity in lung adenocarcinoma progression. Cell. Discov. 9, 96 (2023).
De Zuani, M. et al. Single-cell and Spatial transcriptomics analysis of non-small cell lung cancer. Nat. Commun. 15, 4388 (2024).
Loske, J. et al. Pharmacological improvement of cystic fibrosis transmembrane conductance regulator function rescues airway epithelial homeostasis and host defense in children with cystic fibrosis. Am. J. Respir Crit. Care Med. 209, 1338–1350 (2024).
Januska, M. N. & Walsh, M. J. Single-cell RNA sequencing reveals new basic and translational insights in the cystic fibrosis lung. Am. J. Respir Cell. Mol. Biol. 68, 131–139 (2022).
Cystic Fibrosis Foundation. Cystic Fibrosis Foundation Patient Registry 2022 Annual Data Report (2023).
Wainwright, C. E. et al. Effect of Bronchoalveolar lavage-directed therapy on Pseudomonas aeruginosa infection and structural lung injury in children with cystic fibrosis: a randomized trial. JAMA 306, 163–171 (2011).
Owens, C. M. et al. Lung clearance index and HRCT are complementary markers of lung abnormalities in young children with CF. Thorax 66, 481–488 (2011).
Hisert, K. B. et al. Restoring cystic fibrosis transmembrane conductance regulator function reduces airway bacteria and inflammation in people with cystic fibrosis and chronic lung infections. Am. J. Respir Crit. Care Med. 195, 1617–1628 (2017).
Harris, J. K. et al. Changes in airway Microbiome and inflammation with Ivacaftor treatment in patients with cystic fibrosis and the G551D mutation. Ann. Am. Thorac. Soc. 17, 212–220 (2020).
Zhang, S. et al. Cystic fibrosis macrophage function and clinical outcomes after elexacaftor/tezacaftor/ivacaftor. Eur. Respir J. 61, 2102861 (2023).
Ciobanu, D. Z. et al. Tezacaftor is a direct inhibitor of sphingolipid delta-4 desaturase enzyme (DEGS). J. Cyst. Fibros (2024).
Hao, Y. et al. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat. Biotechnol. 42, 293–304 (2024).
Yang, S. et al. Decontamination of ambient RNA in single-cell RNA-seq with decontx. Genome Biol. 21, 57 (2020).
Jin, S. et al. Inference and analysis of cell-cell communication using cellchat. Nat. Commun. 12, 1088 (2021).
Acknowledgements
We would like to thank our pediatric patients and their families for their generous participation in this study, and we would like to thank the Cystic Fibrosis Center at Lenox Hill Hospital/Northwell Health for graciously entrusting us with the care of their pediatric patients with CF. We would additionally like to thank the Genomics Core Facility and the Mount Sinai Biorepository at the Icahn School of Medicine at Mount Sinai for their essential contributions.
Funding
This work was supported by the Cystic Fibrosis Foundation (JANUSK21D0; awarded to MNJ) as well as the National Center for Advancing Translational Sciences (KL2TR004421; awarded to MNJ) and the National Institute of Diabetes and Digestive and Kidney Diseases (R01DK118946; awarded to MJW) at the National Institutes of Health.
Author information
Authors and Affiliations
Contributions
YS: Methodology, investigation, data curation, formal analysis, visualization, and writing – review and editing; AGV: Writing – review and editing and resources; MBB: Writing – review and editing; MJW: Supervision, methodology, writing – review and editing, funding acquisition, and resources; and MNJ: Conceptualization, methodology, investigation, visualization, writing – original draft, writing – review and editing, funding acquisition, and resources.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Sun, Y., Vicencio, A.G., Beasley, M.B. et al. The single-cell transcriptional landscape of the pediatric cystic fibrosis lung from minimally invasive respiratory specimens. Sci Rep (2026). https://doi.org/10.1038/s41598-026-36125-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-026-36125-w