Abstract
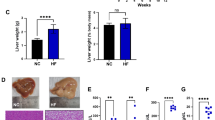
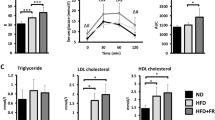
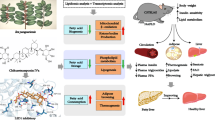
A high-fat diet (HFD) alters the gut microbiota (GM), impairs metabolic efficiency, and increases gut permeability and inflammation. Obesity and insulin resistance are associated with GM dysbiosis. The GM is strongly associated with metabolic disorders and fatty liver disease. The co-chaperone protein FK506-binding protein-5 (FKBP5) regulates several vital cellular processes. Although FKBP5 has been implicated in stress-related disorders, it has not been directly linked to HFD-induced metabolic fatty liver disease. This study aimed to elucidate how FK506 binding protein 5 impairment affects the GM in HFD-induced metabolic dysfunction–associated fatty liver disease and metabolic dysfunction-associated steatotic liver disease (MASLD). Wild-type and FKBP5-knockout (FKKO) mice were fed a normal chow diet or a high-fat diet for 16 weeks. Mouse GM was examined using 16 S rRNA metagenomic analysis. The number of gut-liver immune cells was measured using flow cytometry. HFD-induced hepatic steatosis and inflammation were prevented in FKBP5-deficient mice. FKKO animals showed higher butyric acid levels and GM resistance to diet-induced obesity alterations according to 16 S ribosomal rRNA gene analysis and displayed an HFD-specific gut-liver immunological response that maintained gut barrier failure and mucosal immunity, which are important for GM homeostasis. FKBP5 helps the GM address inadequate immunological responses, including lower gut and liver CD11b+Ly6C+ monocytes and neutrophils, and protects against obesity by improving the GM response to HFD-induced MASLD. FKBP5 protects against HFD-induced MASLD through metabolic coordination between the gut barrier and intrahepatic immunity.
Similar content being viewed by others
Data availability
The data supporting the findings of this study are available from the corresponding author upon request. The sequencing data were deposited in the DNA Data Bank of Japan under accession number PRJNA610239 for 16 S rRNA sequencing.
References
Schmidt, M. V., Paez-Pereda, M., Holsboer, F. & Hausch, F. The prospect of FKBP51 as a drug target. Chem. Med. Chem. 7, 1351–1359. https://doi.org/10.1002/cmdc.201200137 (2012).
Zannas AS, Wiechmann T, Gassen NC, Binder EB. Gene-Stress-Epigenetic Regulation of FKBP5: Clinical and Translational Implications. Neuropsychopharmacology 41(1):261-74. https://doi.org/10.1038/npp.2015.235 (2016).
Balsevich, G. et al. Stress-responsive FKBP51 regulates AKT2-AS160 signaling and metabolic function. Nat. Commun. 8, 1725. https://doi.org/10.1038/s41467-017-01783-y (2017).
Feng, B. et al. Mitogen-activated protein kinase phosphatase 3 (MKP-3)-deficient mice are resistant to diet-induced obesity. Diabetes 63, 2924–2934. https://doi.org/10.2337/db14-0066 (2014).
Häusl, A. S. et al. Mediobasal hypothalamic FKBP51 acts as a molecular switch linking autophagy to whole-body metabolism. Sci. Adv. 8, 4797. https://doi.org/10.1126/sciadv.abi4797 (2022).
Stechschulte, L. A. et al. FKBP51 null mice are resistant to diet-induced obesity and the PPARgamma agonist Rosiglitazone. Endocrinology 157, 3888–3900. https://doi.org/10.1210/en.2015-1996 (2016).
Yang, L. et al. Hypothalamic Fkbp51 is induced by fasting, and elevated hypothalamic expression promotes obese phenotypes. Am. J. Physiol. Endocrinol. Metab. 302, E987–E991. https://doi.org/10.1152/ajpendo.00474.2011 (2012).
Xu, H. et al. Protective effect of Lactobacillus rhamnosus GG and its supernatant against myocardial dysfunction in obese mice exposed to intermittent hypoxia is associated with the activation of Nrf2 pathway. Int. J. Biol. Sci. 15, 2471–2483. https://doi.org/10.7150/ijbs.36465 (2019).
Vallianou, N., Stratigou, T., Christodoulatos, G. S. & Dalamaga, M. Understanding the role of the gut Microbiome and microbial metabolites in obesity and obesity-associated metabolic disorders: current evidence and perspectives. Curr. Obes. Rep. 8, 317–332. https://doi.org/10.1007/s13679-019-00352-2 (2019).
Safari, Z. & Gérard, P. The links between the gut Microbiome and non-alcoholic fatty liver disease (NAFLD). Cell. Mol. Life Sci. 76., 1541–1558. https://doi.org/10.1007/s00018-019-03011-w (2019).
Qiu, P. et al. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat. Biotechnol. 29, 886–891. https://doi.org/10.1038/nbt.1991 (2011).
Kim, B., Choi, H. N. & Yim, J. E. Effect of diet on the gut microbiota associated with obesity. J. Obes. Metab. Syndr. 28, 216–224. https://doi.org/10.7570/jomes.2019.28.4.216 (2019).
Lin YN, Hsu JR, Wang CL, Huang YC, Wang JY, Wu CY, Wu LL. Nuclear factor interleukin 3 and metabolic dysfunction-associated fatty liver disease development. Commun Biol. 2024 Jul 24;7(1):897. doi: 10.1038/s42003-024-06565-z.
Murphy, E. F. et al. Divergent metabolic outcomes arising from targeted manipulation of the gut microbiota in diet-induced obesity. Gut 62, 220–226. https://doi.org/10.1136/gutjnl-2011-300705 (2013).
Boursier, J. & Diehl, A. M. Implication of gut microbiota in nonalcoholic fatty liver disease. PLOS Pathog. 11, e1004559. https://doi.org/10.1371/journal.ppat.1004559 (2015).
Turnbaugh, P. J. et al. An obesity-associated gut Microbiome with increased capacity for energy harvest. Nature 444, 1027–1031. https://doi.org/10.1038/nature05414 (2006).
Huang, E. Y. et al. Composition of dietary fat source shapes gut microbiota architecture and alters host inflammatory mediators in mouse adipose tissue. JPEN J. Parenter. Enter. Nutr. 37, 746–754. https://doi.org/10.1177/0148607113486931 (2013).
Tong, X. et al. Structural alteration of gut microbiota during the amelioration of human type 2 diabetes with hyperlipidemia by Metformin and a traditional Chinese herbal formula: a multicenter, randomized, open label clinical trial. mBio 9, e02392–e02317. https://doi.org/10.1128/mBio.02392-17 (2018).
Murri, M. et al. Gut microbiota in children with type 1 diabetes differs from that in healthy children: a case-control study. BMC Med. 11, 46. https://doi.org/10.1186/1741-7015-11-46 (2013).
Liu, X. et al. Blautia-a new functional genus with potential probiotic properties? Gut Microbes. 13, 1–21. https://doi.org/10.1080/19490976.2021.1875796 (2021).
González-Navajas, J. M. et al. TLR4 signaling in effector CD4 + T cells regulates TCR activation and experimental colitis in mice. J. Clin. Invest. 120, 570–581. https://doi.org/10.1172/JCI40055 (2010).
Zannas, A. S. et al. Epigenetic upregulation of FKBP5 by aging and stress contributes to NF-κB-driven inflammation and cardiovascular risk. Proc. Natl. Acad. Sci. U. S. A. . 116, 11370–11379 (2019). https://doi.org/10.1073/pnas.1816847116
Lee, S. et al. Blueberry supplementation influences the gut microbiota, inflammation, and insulin resistance in high-fat-diet-fed rats. J. Nutr. 148, 209–219. https://doi.org/10.1093/jn/nxx027 (2018).
Zhou, X. L., Yan, B. B., Xiao, Y., Zhou, Y. M. & Liu, T. Y. Tartary buckwheat protein prevented dyslipidemia in high-fat diet-fed mice associated with gut microbiota changes. Food Chem. Toxicol. 119, 296–301. https://doi.org/10.1016/j.fct.2018.02.052 (2018).
Cani, P. D. et al. Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP-2-driven improvement of gut permeability. Gut 58, 1091–1103. https://doi.org/10.1136/gut.2008.165886 (2009).
Li, M. et al. Gut carbohydrate metabolism instead of fat metabolism regulated by gut microbes mediates high-fat diet-induced obesity. Benef Microbes. 5, 335–344. https://doi.org/10.3920/BM2013.0071 (2014).
Alcock, J. & Lin, H. C. Fatty acids from diet and microbiota regulate energy metabolism. F1000Res 4, 738. https://doi.org/10.12688/f1000research.6078.1 (2015).
Ghaleb, A. M., McConnell, B. B., Kaestner, K. H. & Yang, V. W. Altered intestinal epithelial homeostasis in mice with intestine-specific deletion of the Kruppel-like factor 4 gene. Dev. Biol. 349, 310–320. https://doi.org/10.1016/j.ydbio.2010.11.001 (2011).
Li, H. et al. Crosstalk between liver macrophages and surrounding cells in nonalcoholic steatohepatitis. Front. Immunol. 11, 1169. https://doi.org/10.3389/fimmu.2020.01169 (2020).
Wang, X. et al. STING expression in monocyte-derived macrophages is associated with the progression of liver inflammation and fibrosis in patients with nonalcoholic fatty liver disease. Lab. Invest. 100, 542–552. https://doi.org/10.1038/s41374-019-0342-6 (2020).
Acknowledgements
We would like to express our gratitude to Dr. Yu-Lun Kuo of BIOTOOLS Co., Ltd., Taiwan, for assisting us with the NGS data analysis. We would also like to express our gratitude for the technical help offered by the flow cytometric analysis, sorting, and imaging cores of the First Core Laboratory at the National Taiwan and Yang Ming University College of Medicine. We thank the NYCU Microbiota Research Center and the Taiwan Mouse Clinical National Comprehensive Mouse Phenotyping and Drug Testing Center. We thank National Center for Biomodels (NCB), NIAR, Taiwan, for technical support in service of isolator. We thank the Taiwan Mouse Clinic, Academia Sinica, and the Taiwan Animal Consortium for their technical support. NLAC (NARLabs, Taiwan) provided technical assistance to isolators. The authors acknowledge the research collaboration and technical service supported by National Human Microbiome Core Facility, Taiwan. We acknowledge the research collaboration and technical service supported by National Human Microbiome Core Facility. FKBP5 KO mice were obtained from Dr. Yi Hsuan Lee. We are grateful to the summer student Zi-Yun Wang (Department of Pharmacology & Toxicology, University of Toronto, Toronto, ON M5S 1A8, Canada) for assistance with data analysis. All authors discussed the results and approved the manuscript.
Funding
Ministry of Science and Technology (MOST) of Taiwan, Grant Number: 108-2320-B-010-045-MY3, 110-2320-B-002-080-MY3, MOST 111-2314-B-A49-072, NSTC 112-2314-B-A49-028-MY3, NSTC 112-2740-B-A49-002, NSTC 113-2740-B-A49-003, NSTC 113-2321-B-A49-014-, NSTC 114-2321-B-A49-004 -, NSTC 114-2740-B-A49-003- (to LLW), and MOST 106-2320-B-010-009-MY3 (to CCJ); Yen Tjing Ling Medical Foundation, Grant Number: CI-110-22,CI-111-24 and CI-115-33 (to LLW).
Author information
Authors and Affiliations
Contributions
Li-Ling Wu conceived and designed the experiments; Yu-Jen Liao, Luen-Kui Chen, Yi-Chen Huang, Chia-Yen Chen, Wei-Hao Peng Performed the experiments; Li-Ling Wu and Yu-Jen Liao conducted the data analysis; Li-Ling Wu and Chi-Chang Juan revised the manuscript; Li-Ling Wu Wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wu, LL., Liao, YJ., Peng, WH. et al. FK506-binding protein-5 in high-fat diet-induced metabolic dysfunction-associated steatotic liver disease. Sci Rep (2026). https://doi.org/10.1038/s41598-026-38549-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-026-38549-w