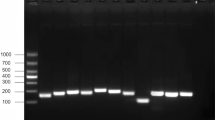
Abstract
Food-borne pathogens represent a significant threat to global food safety and public health. Enterohemorrhagic Escherichia coli (EHEC) O157:H7, Salmonella, and Cronobacter are among the principal bacterial agents responsible for food-borne illnesses. Advances in sequencing technology have made it possible to identify particular genetic markers for these pathogens through large-scale genomic analysis. In this study, two conserved genes, z0340 and sbcC, were screened in silico from extensive bacterial genomes (including 1,460,756 genomes) for EHEC O157:H7 and Salmonella, respectively. Based on these two newly identified markers, combined with previously identified Cronobacter marker gene ygcB, multiplex PCR and multiplex TaqMan quantitative PCR methods were developed and established for the simultaneous detection of EHEC O157:H7, Salmonella, and Cronobacter. The specificity of the multiplex PCR and multiplex TaqMan qPCR was determined using 15 strains of Cronobacter, one strain of EHEC O157:H7, seven strains of Salmonella, and 100 strains of other common environmental bacteria and pathogens. The results showed that two methods demonstrated 100% specificity, both for multiple PCR and TaqMan qPCR. The detection limits of the multiplex PCR and TaqMan qPCR assays were 1 pg/μL and 0.5 pg/μL, respectively. Therefore, the newly developed methods are sensitive and reliable for the simultaneous identification of the three common food-borne pathogens.
Similar content being viewed by others
Data availability
All data supporting the findings of this study are available within the paper and its Supplementary Information. Furthermore, the original datasets (including in-house scripts) generated and/or analyzed during the current study are publicly available in the detect_rawdata repository, accessible via the GitHub platform: https://github.com/happywlu/detect_rawdata.
References
Allard, M. W. et al. Genomics of food-borne pathogens for microbial food safety. Curr. Opin. Biotechnol. 49(2), 224–229. https://doi.org/10.1016/j.copbio.2017.11.002 (2018).
Deng, L. & Wang, S. Colonization resistance: The role of gut microbiota in preventing Salmonella invasion and infection. Gut Microbes 16(1), 2424914. https://doi.org/10.1080/19490976 (2024).
Sun, H. et al. Regulation of flagellar motility and biosynthesis in Enterohemorrhagic Escherichia coli O157:H7. Gut Microbes 14(1), 2110822. https://doi.org/10.1080/19490976 (2022).
Haston, J. C. et al. Cronobacter sakazakii infections in two infants linked to powdered infant formula and breast pump equipment-united states, 2021 and 2022. Mmwr. Morb. Mortal. Wkly. Rep. 72(9), 223–226. https://doi.org/10.15585/mmwr.mm7209a2 (2023).
Withenshaw, S. M. et al. A systematized review and qualitative synthesis of potential risk factors associated with the occurrence of non-O157 shiga toxin-producing Escherichia coli (STEC) in the primary production of cattle. Compr. Rev. Food Sci. Food Saf. 21(3), 2363–2390. https://doi.org/10.1111/1541-4337 (2022).
Lu, J. et al. Salmonella: Infection mechanism and control strategies. Microbiol. Res. 292, 128013. https://doi.org/10.1016/j.micres (2025).
Strysko, J. et al. Food safety and invasive Cronobacter infections during early infancy, 1961–2018. Emerg. Infect. Dis. 26(5), 857–865. https://doi.org/10.3201/eid2605 (2020).
Farhana, A. & Khan, Y. S. Biochemistry (Statpearls Publishing, 2023).
Zhang, S. et al. Characterization of Escherichia coli O157:Non-H7 isolated from retail food in China and first report of mcr-1/inci2-carrying colistin-resistant E. coli O157:H26 and E. coli O157:H4. Int. J. Food Microbiol. 378, 109805. https://doi.org/10.1016/j.ijfoodmicro (2022).
Fierer, J. Invasive non-typhoidal Salmonella (Ints) infections. Clin. Infect. Dis. 75(4), 732–738. https://doi.org/10.1093/cid/ciac035 (2022).
Ruddle, S. J. et al. Salmonella-Liberated dietary l-arabinose promotes expansion in superspreaders. Cell Host Microbe 31(3), 405–417. https://doi.org/10.1016/j.chom (2023).
Jiang, L. et al. Virulence-related O islands in enterohemorrhagic Escherichia coli O157:H7. Gut Microbes 13(1), 1992237. https://doi.org/10.1080/19490976 (2021).
Fu, H. et al. Rapid detection of goose megrivirus using TaqMan real-time PCR technology. Poult. Sci. 103(5), 103611. https://doi.org/10.1016/j.psj (2024).
Ndraha, N., Lin, H.-Y., Wang, C.-Y., Hsiao, H.-I. & Lin, H.-J. Rapid detection methods for foodborne pathogens based on nucleic acid amplification: Recent advances, remaining challenges, and possible opportunities. Food Chem. Mol. Sci. 7, 100183. https://doi.org/10.1016/j.fochms.2023.100183 (2023).
Barton, A. A. et al. Host restriction, pathogenesis and chronic carriage of typhoidal Salmonella. Fems Microbiol. Rev. 45(5), 41. https://doi.org/10.1093/femsre/fuab014 (2021).
Hazen, T. H. et al. Genomic diversity of EPEC associated with clinical presentations of differing severity. Nat. Microbiol. 1(18), 15014. https://doi.org/10.1038/nmicrobiol.2015.14 (2016).
Juscamayta-López, E. V. F. S. M. A pangenome approach-based loop-mediated isothermal amplification assay for the specific and early detection of Bordetella Pertussis. Sci. Rep. 1(13), 4356. https://doi.org/10.1038/s41598-023-29773-9 (2023).
Wang, L. et al. Detection of genus and three important species of Cronobacter using novel genus-and species-specific genes identified by large-scale comparative genomic analysis. Front. Microbiol. 13(13), 885543. https://doi.org/10.3389/fmicb.2022.885543 (2022).
Pava-Ripoll, M. et al. Ingested Salmonella Enterica, Cronobacter sakazakii, Escherichia coli O157:H7, and listeria monocytogenes: transmission dynamics from adult house flies to their eggs and first filial (F1) generation adults. BMC Microbiol. 15(15), 150. https://doi.org/10.1186/s12866-015-0478-5 (2015).
Jk, J. Marker and reporter genes: Illuminating tools for environmental microbiologists. Curr. Opin. Microbiol. 3(6), 310–316. https://doi.org/10.1016/s1369-5274(03)00057-2 (2003).
Meng, H. et al. High-throughput host-microbe single-cell RNA sequencing reveals ferroptosis-associated heterogeneity during Acinetobacter baumannii infection. Angew. Chem. Int. Ed. Engl. 63(18), e202400538. https://doi.org/10.1002/anie.202400538 (2024).
Qin, X. et al. Comparative genomics reveals environmental adaptation differences between Cronobacter species. Food Res. Int. 147, 110541. https://doi.org/10.1016/j.foodres.2021.110541 (2021).
Papalia, M. et al. Genomic characterization of Achromobacter genogroup 20 and identification of a potential species-specific marker. J. Glob. Antimicrob. Resist. 40, 26–28. https://doi.org/10.1016/j.jgar.2024.11.012 (2025).
Massetti, L. et al. A Taq-Man-based multiplex quantitative PCR for the simultaneous detection and quantification of angiostrongylus vasorum, crenosoma vulpis, and species of respiratory capillarids in canids. Int. J. Parasitol. 54(3–4), 185–193. https://doi.org/10.1016/j.ijpara.2023.12.001 (2024).
Lee, J. I., Kim, S. S. & Kang, D. H. Development of DNA probes to detect Cronobacter Sakazakii based on comparative genomics and its application in food samples. Food Control 137, 108853 (2022).
Goay Yx, C. K. T. C. Identification of five novel Salmonella typhi-specific genes as markers for diagnosis of typhoid fever using single-gene target PCR assays. Biomed. Res. Int. 2016, 8905675. https://doi.org/10.1155/2016/8905675 (2016).
Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 30(14), 2068–2069. https://doi.org/10.1093/bioinformatics/btu153 (2014).
Page, A. J. et al. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 31(22), 3691–3693. https://doi.org/10.1093/bioinformatics/btv421 (2015).
Bessonov, K. et al. Ectyper: In silico Escherichia coli serotype and species prediction from raw and assembled whole-genome sequence data. Microb. Genom. 7(12), 000728. https://doi.org/10.1099/mgen.0.000728 (2021).
Sahl, J. W. et al. The large-scale blast score ratio (ls-bsr) pipeline: A method to rapidly compare genetic content between bacterial genomes. PeerJ 2, e332. https://doi.org/10.7717/peerj.332 (2014).
Premier Biosoft. Primer Premier 6 [cited 2024 Date]. Available from: https://www.premierbiosoft.com/primerdesign/ (2024).
Integrated DNA Technologies. OligoAnalyzer Tool [Internet] [cited 2024 Date]. Available from: https://www.idtdna.com/pages/tools/oligoanalyzer (2024).
Kobayashi, H. et al. Simultaneous enrichment of Salmonella spp., Escherichia coli O157:H7, Vibrio parahaemolyticus, Staphylococcus aureus, Bacillus cereus, and Listeria monocytogenes by single broth and screening of the pathogens by multiplex real-time PCR. Food Sci. Technol. Res. 15(4), 427–438 (2009).
Nguyen, T. T., Van Giau, V. & Vo, T. K. Multiplex PCR for simultaneous identification of E. coli O157:H7, Salmonella spp. and L. monocytogenes in food. 3 Biotech 6, 205. https://doi.org/10.1007/s13205-016-0523-6 (2016).
Boukharouba, A., González, A., García-Ferrús, M., Ferrús, M. A. & Botella, S. Simultaneous detection of four main foodborne pathogens in ready-to-eat food by using a simple and rapid multiplex PCR (mPCR) assay. Int. J. Environ. Res. Public Health 19, 1031. https://doi.org/10.3390/ijerph19031031 (2022).
Altschul Sf, G. W. M. W. Basic local alignment search tool. J. Mol. Biol. 3(215), 403–410. https://doi.org/10.1016/S0022-2836(05)80360-2 (1990).
Lee, N. et al. A Multiplex PCR assay for simultaneous detection of Escherichia coli O157:H7, Bacillus cereus, Vibrio parahaemolyticus, Salmonella spp., Listeria monocytogenes, and Staphylococcus aureus in Korean ready-to-eat food. Foodborne Pathog. Dis. 11, 574–580. https://doi.org/10.1089/fpd.2013.1638 (2014).
Funding
This study is supported by the Natural Science Foundation of China (32201986), Natural Science Foundation of Hubei Province (2022CFB743), and Open Fund of Hubei Provincial Clinical Research Center for Precise Prevention and Treatment of Elderly Gastrointestinal Cancer (2024EGC-01).
Author information
Authors and Affiliations
Contributions
L. W. and L. Y. designed the research. H.N. Z., P. X., Z. L., and performed the research. X.L. C., R.H. Y., and L.L. Z. collected the data. Q.Y. Z., T.Y. L., and X.W. T. provided technical support and insights. P. X., Z. L., and L. W. analyzed the data. H.N. Z., P. X., and L. W. wrote the manuscript. L. W. revised the manuscript. All authors contributed to the article and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The conducted research is not related to either human or animal use.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, H., Xiong, P., Lu, Z. et al. Novel marker genes for simultaneous detection of Salmonella, EHEC O157:H7, and Cronobacter. Sci Rep (2026). https://doi.org/10.1038/s41598-026-38990-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-026-38990-x